library(BayesMallows)
set.seed(123)This vignette describes how to run Markov chain Monte Carlo with parallel chains. For an introduction to the “BayesMallows” package, please see the introductory vignette, which is an updated version of Sørensen et al. (2020). For parallel processing of particles with the sequential Monte Carlo algorithm of Stein (2023), see the SMC vignette.
Why Parallel Chains?
Modern computers have multiple cores, and on computing clusters one can get access to hundreds of cores easily. By running Markov Chains in parallel on cores, ideally from different starting points, we achieve at least the following:
The time you have to wait to get the required number of post-burnin samples scales like .
You can check convergence by comparing chains.
Parallel Chains with Complete Rankings
In “BayesMallows” we use the “parallel” package for parallel computation. Parallelization is obtained by starting a cluster and providing it as an argument. Note that we also give one initial value of the dispersion parameter to each chain.
library(parallel)
cl <- makeCluster(4)
fit <- compute_mallows(
data = setup_rank_data(rankings = potato_visual),
compute_options = set_compute_options(nmc = 5000),
cl = cl
)
stopCluster(cl)We can assess convergence in the usual way:
assess_convergence(fit)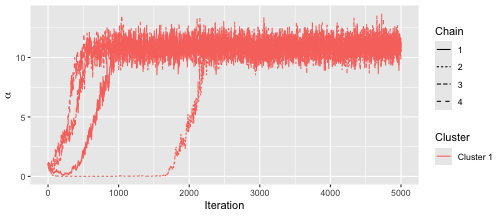
We can also assess convergence for the latent ranks . Since the initial value of is sampled uniformly, the two chains automatically get different initial values.
assess_convergence(fit, parameter = "rho", items = 1:3)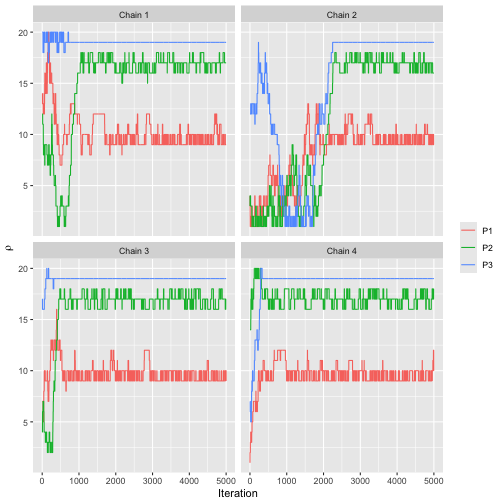
Based on the convergence plots, we set the burnin to 3000.
burnin(fit) <- 3000We can now use all the tools for assessing the posterior distributions as usual. The post-burnin samples for all parallel chains are simply combined, as they should be.
Below is a plot of the posterior distribution of .
plot(fit)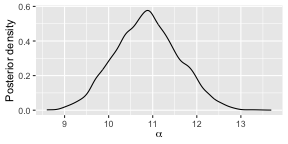
Next is a plot of the posterior distribution of .
plot(fit, parameter = "rho", items = 4:7)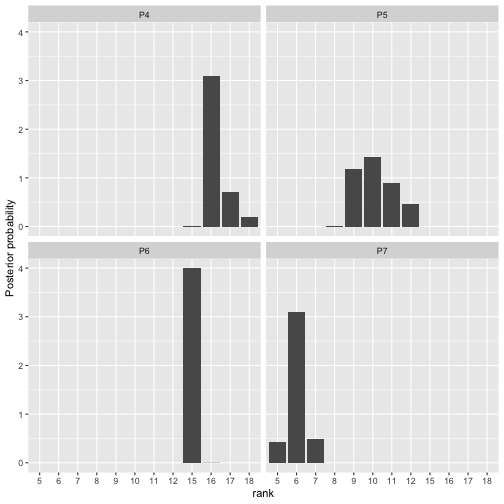
Parallel Chains with Pairwise Preferences
A case where parallel chains might be more strongly needed is with incomplete data, e.g., arising from pairwise preferences. In this case the MCMC algorithm needs to perform data augmentation, which tends to be both slow and sticky. We illustrate this with the beach preference data, again referring to Sørensen et al. (2020) for a more thorough introduction to the aspects not directly related to parallelism.
beach_data <- setup_rank_data(preferences = beach_preferences)We run four parallel chains, letting the package generate random initial rankings, but again providing a vector of initial values for .
cl <- makeCluster(4)
fit <- compute_mallows(
data = beach_data,
compute_options = set_compute_options(nmc = 4000, save_aug = TRUE),
initial_values = set_initial_values(alpha_init = runif(4, 1, 4)),
cl = cl
)
stopCluster(cl)Trace Plots
The convergence plots shows some long-range autocorrelation, but otherwise it seems to mix relatively well.
assess_convergence(fit)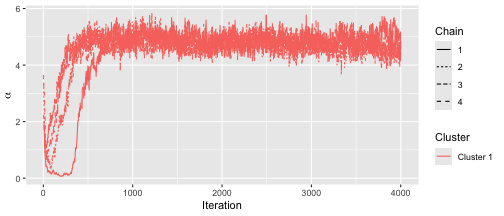
Here is the convergence plot for :
assess_convergence(fit, parameter = "rho", items = 4:6)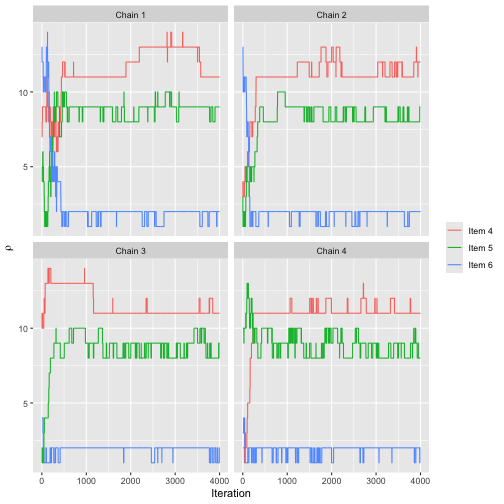
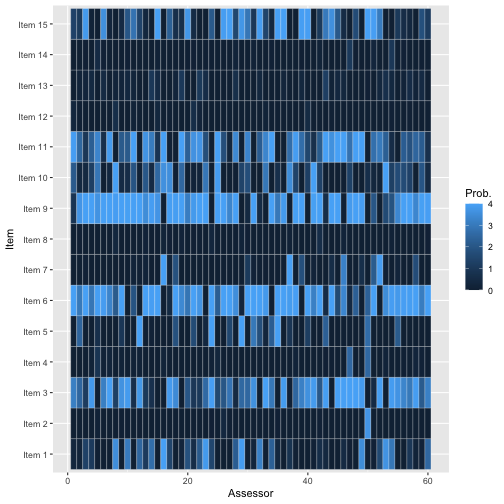
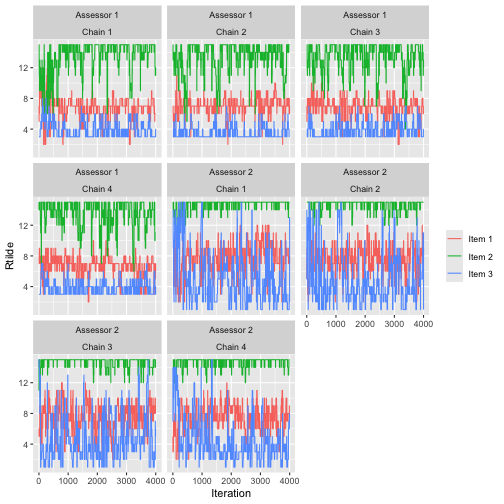
To avoid overplotting, it’s a good idea to pick a low number of assessors and chains. We here look at items 1-3 of assessors 1 and 2.
assess_convergence(fit,
parameter = "Rtilde",
items = 1:3, assessors = 1:2
)
Posterior Quantities
Based on the trace plots, the chains seem to be mixing well. We set the burnin to 1000.
burnin(fit) <- 1000We can now study the posterior distributions. Here is the posterior
for
.
Note that by increasing the nmc argument to
compute_mallows above, the density would appear smoother.
In this vignette we have kept it low to reduce the run time.
plot(fit)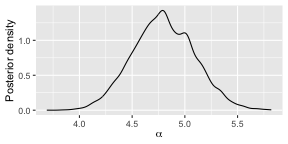
We can also look at the posterior for .
plot(fit, parameter = "rho", items = 6:9)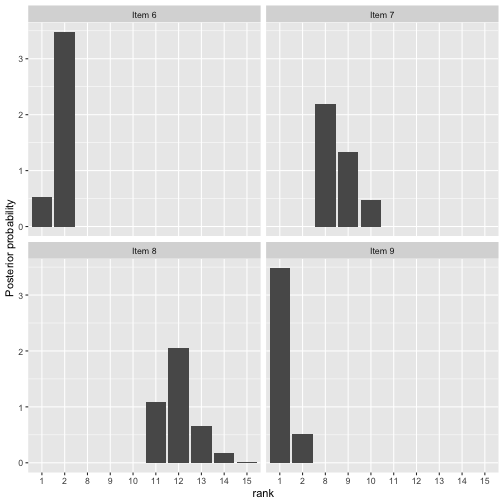
We can also compute posterior intervals in the usual way:
compute_posterior_intervals(fit, parameter = "alpha")
#> parameter mean median hpdi central_interval
#> 1 alpha 4.798 4.793 [4.242,5.373] [4.235,5.371]
compute_posterior_intervals(fit, parameter = "rho")
#> parameter item mean median hpdi central_interval
#> 1 rho Item 1 7 7 [7] [6,7]
#> 2 rho Item 2 15 15 [15] [14,15]
#> 3 rho Item 3 3 3 [3,4] [3,4]
#> 4 rho Item 4 11 11 [11,13] [11,13]
#> 5 rho Item 5 9 9 [8,10] [8,10]
#> 6 rho Item 6 2 2 [1,2] [1,2]
#> 7 rho Item 7 9 8 [8,10] [8,10]
#> 8 rho Item 8 12 12 [11,13] [11,14]
#> 9 rho Item 9 1 1 [1,2] [1,2]
#> 10 rho Item 10 6 6 [5,6] [5,7]
#> 11 rho Item 11 4 4 [3,5] [3,5]
#> 12 rho Item 12 13 13 [12,14] [11,14]
#> 13 rho Item 13 10 10 [8,10] [8,10]
#> 14 rho Item 14 13 14 [12,14] [12,14]
#> 15 rho Item 15 5 5 [4,5] [4,6]And we can compute the consensus ranking:
compute_consensus(fit)
#> cluster ranking item cumprob
#> 1 Cluster 1 1 Item 9 0.8691667
#> 2 Cluster 1 2 Item 6 1.0000000
#> 3 Cluster 1 3 Item 3 0.6391667
#> 4 Cluster 1 4 Item 11 0.9404167
#> 5 Cluster 1 5 Item 15 0.9559167
#> 6 Cluster 1 6 Item 10 0.9636667
#> 7 Cluster 1 7 Item 1 1.0000000
#> 8 Cluster 1 8 Item 7 0.5473333
#> 9 Cluster 1 9 Item 5 0.9255833
#> 10 Cluster 1 10 Item 13 1.0000000
#> 11 Cluster 1 11 Item 4 0.6924167
#> 12 Cluster 1 12 Item 8 0.7833333
#> 13 Cluster 1 13 Item 12 0.6158333
#> 14 Cluster 1 14 Item 14 0.9958333
#> 15 Cluster 1 15 Item 2 1.0000000
compute_consensus(fit, type = "MAP")
#> cluster map_ranking item probability
#> 1 Cluster 1 1 Item 9 0.2683333
#> 2 Cluster 1 2 Item 6 0.2683333
#> 3 Cluster 1 3 Item 3 0.2683333
#> 4 Cluster 1 4 Item 11 0.2683333
#> 5 Cluster 1 5 Item 15 0.2683333
#> 6 Cluster 1 6 Item 10 0.2683333
#> 7 Cluster 1 7 Item 1 0.2683333
#> 8 Cluster 1 8 Item 7 0.2683333
#> 9 Cluster 1 9 Item 5 0.2683333
#> 10 Cluster 1 10 Item 13 0.2683333
#> 11 Cluster 1 11 Item 4 0.2683333
#> 12 Cluster 1 12 Item 8 0.2683333
#> 13 Cluster 1 13 Item 12 0.2683333
#> 14 Cluster 1 14 Item 14 0.2683333
#> 15 Cluster 1 15 Item 2 0.2683333We can compute the probability of being top-, here for :
plot_top_k(fit, k = 4)