Compute the posterior distributions of the parameters of the Bayesian Mallows Rank Model, given rankings or preferences stated by a set of assessors.
The BayesMallows package uses the following parametrization of the
Mallows rank model (Mallows 1957)
:
$$p(r|\alpha,\rho) = \frac{1}{Z_{n}(\alpha)} \exp\left\{\frac{-\alpha}{n} d(r,\rho)\right\}$$
where \(r\) is a ranking, \(\alpha\) is a scale parameter, \(\rho\) is the latent consensus ranking, \(Z_{n}(\alpha)\) is the partition function (normalizing constant), and \(d(r,\rho)\) is a distance function measuring the distance between \(r\) and \(\rho\). We refer to Vitelli et al. (2018) for further details of the Bayesian Mallows model.
compute_mallows always returns posterior distributions of the latent
consensus ranking \(\rho\) and the scale parameter \(\alpha\). Several
distance measures are supported, and the preferences can take the form of
complete or incomplete rankings, as well as pairwise preferences.
compute_mallows can also compute mixtures of Mallows models, for
clustering of assessors with similar preferences.
Usage
compute_mallows(
data,
model_options = set_model_options(),
compute_options = set_compute_options(),
priors = set_priors(),
initial_values = set_initial_values(),
pfun_estimate = NULL,
progress_report = set_progress_report(),
cl = NULL
)Arguments
- data
An object of class "BayesMallowsData" returned from
setup_rank_data().- model_options
An object of class "BayesMallowsModelOptions" returned from
set_model_options().- compute_options
An object of class "BayesMallowsComputeOptions" returned from
set_compute_options().- priors
An object of class "BayesMallowsPriors" returned from
set_priors().- initial_values
An object of class "BayesMallowsInitialValues" returned from
set_initial_values().- pfun_estimate
Object returned from
estimate_partition_function(). Defaults toNULL, and will only be used for footrule, Spearman, or Ulam distances when the cardinalities are not available, cf.get_cardinalities().- progress_report
An object of class "BayesMallowsProgressReported" returned from
set_progress_report().- cl
Optional cluster returned from
parallel::makeCluster(). If provided, chains will be run in parallel, one on each node ofcl.
References
Mallows CL (1957).
“Non-Null Ranking Models. I.”
Biometrika, 44(1/2), 114–130.
Vitelli V, Sørensen, Crispino M, Arjas E, Frigessi A (2018).
“Probabilistic Preference Learning with the Mallows Rank Model.”
Journal of Machine Learning Research, 18(1), 1–49.
https://jmlr.org/papers/v18/15-481.html.
See also
Other modeling:
burnin(),
burnin<-(),
compute_mallows_mixtures(),
compute_mallows_sequentially(),
sample_prior(),
update_mallows()
Examples
# ANALYSIS OF COMPLETE RANKINGS
# The example datasets potato_visual and potato_weighing contain complete
# rankings of 20 items, by 12 assessors. We first analyse these using the Mallows
# model:
set.seed(1)
model_fit <- compute_mallows(
data = setup_rank_data(rankings = potato_visual),
compute_options = set_compute_options(nmc = 2000)
)
# We study the trace plot of the parameters
assess_convergence(model_fit, parameter = "alpha")
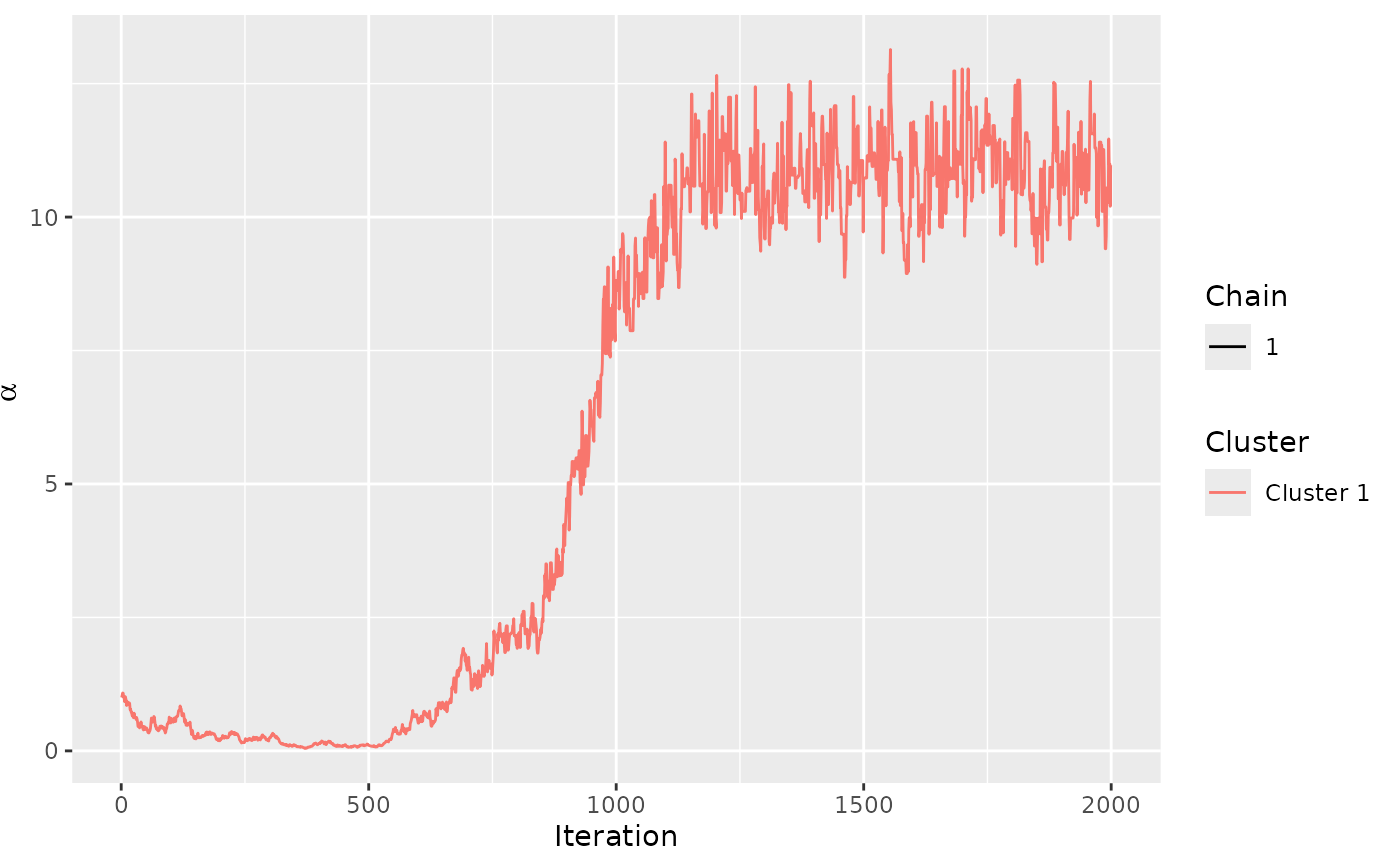 assess_convergence(model_fit, parameter = "rho", items = 1:4)
assess_convergence(model_fit, parameter = "rho", items = 1:4)
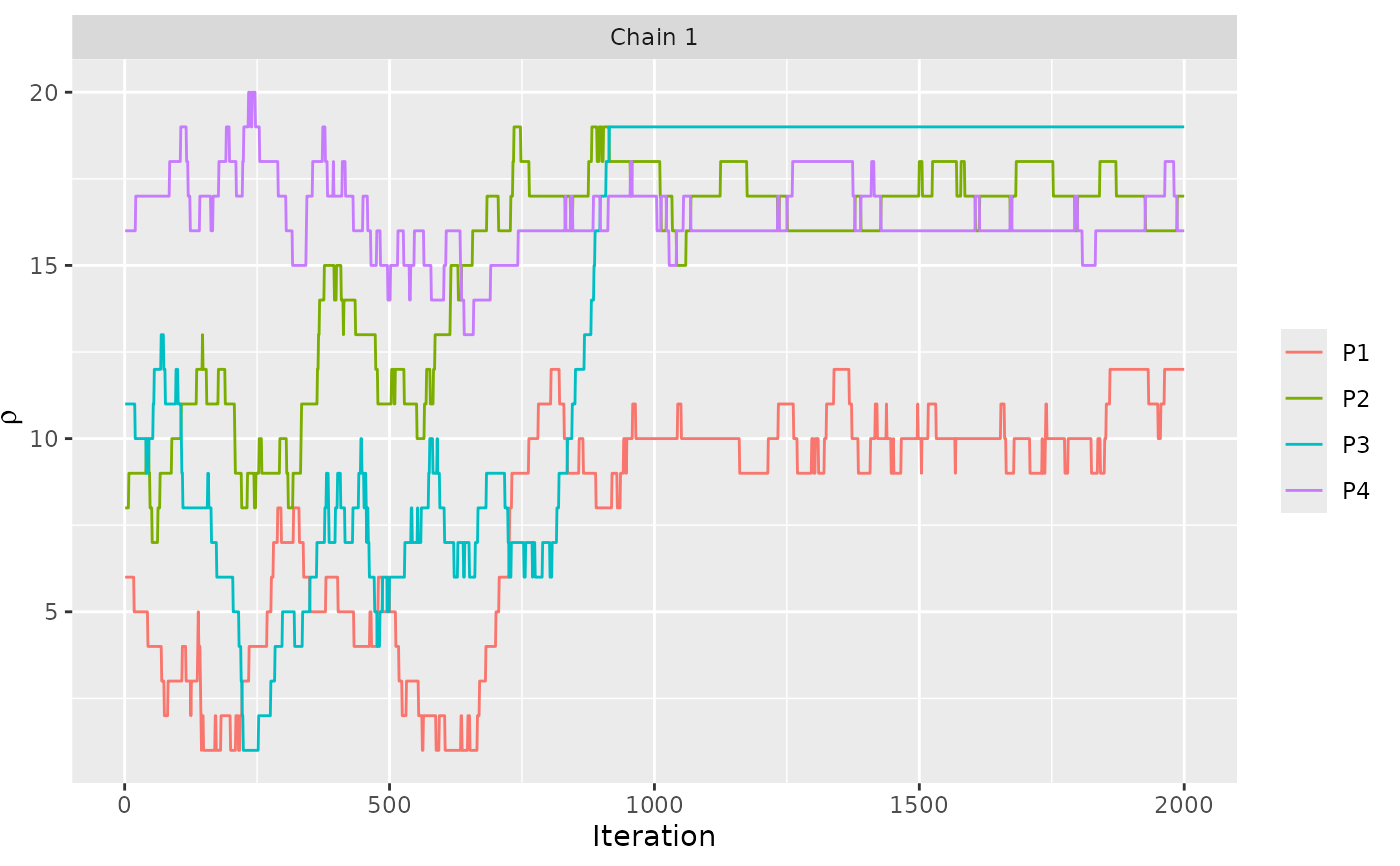 # Based on these plots, we set burnin = 1000.
burnin(model_fit) <- 1000
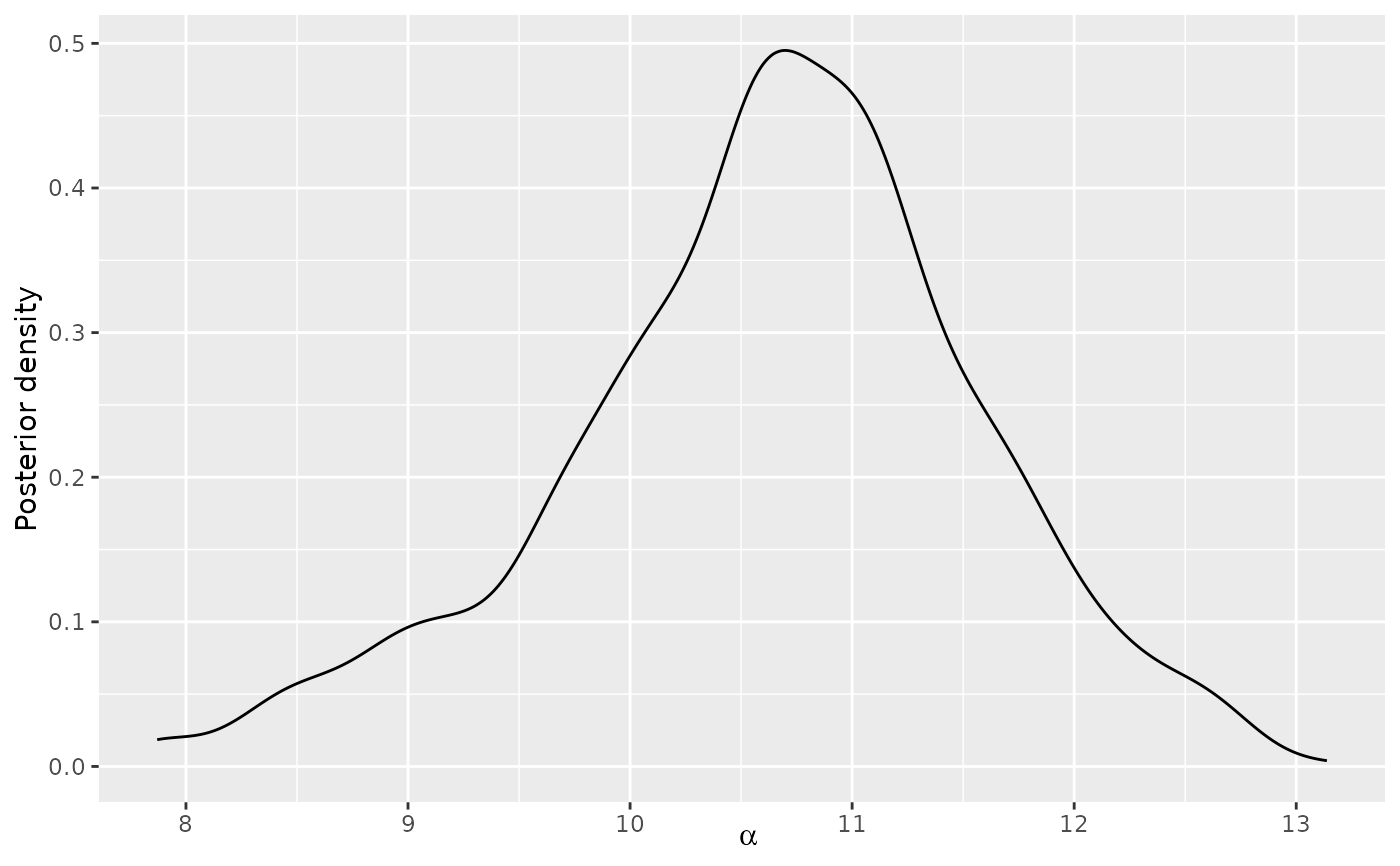
# Next, we use the generic plot function to study the posterior distributions
# of alpha and rho
plot(model_fit, parameter = "alpha")
# Based on these plots, we set burnin = 1000.
burnin(model_fit) <- 1000
# Next, we use the generic plot function to study the posterior distributions
# of alpha and rho
plot(model_fit, parameter = "alpha")
 plot(model_fit, parameter = "rho", items = 10:15)
plot(model_fit, parameter = "rho", items = 10:15)
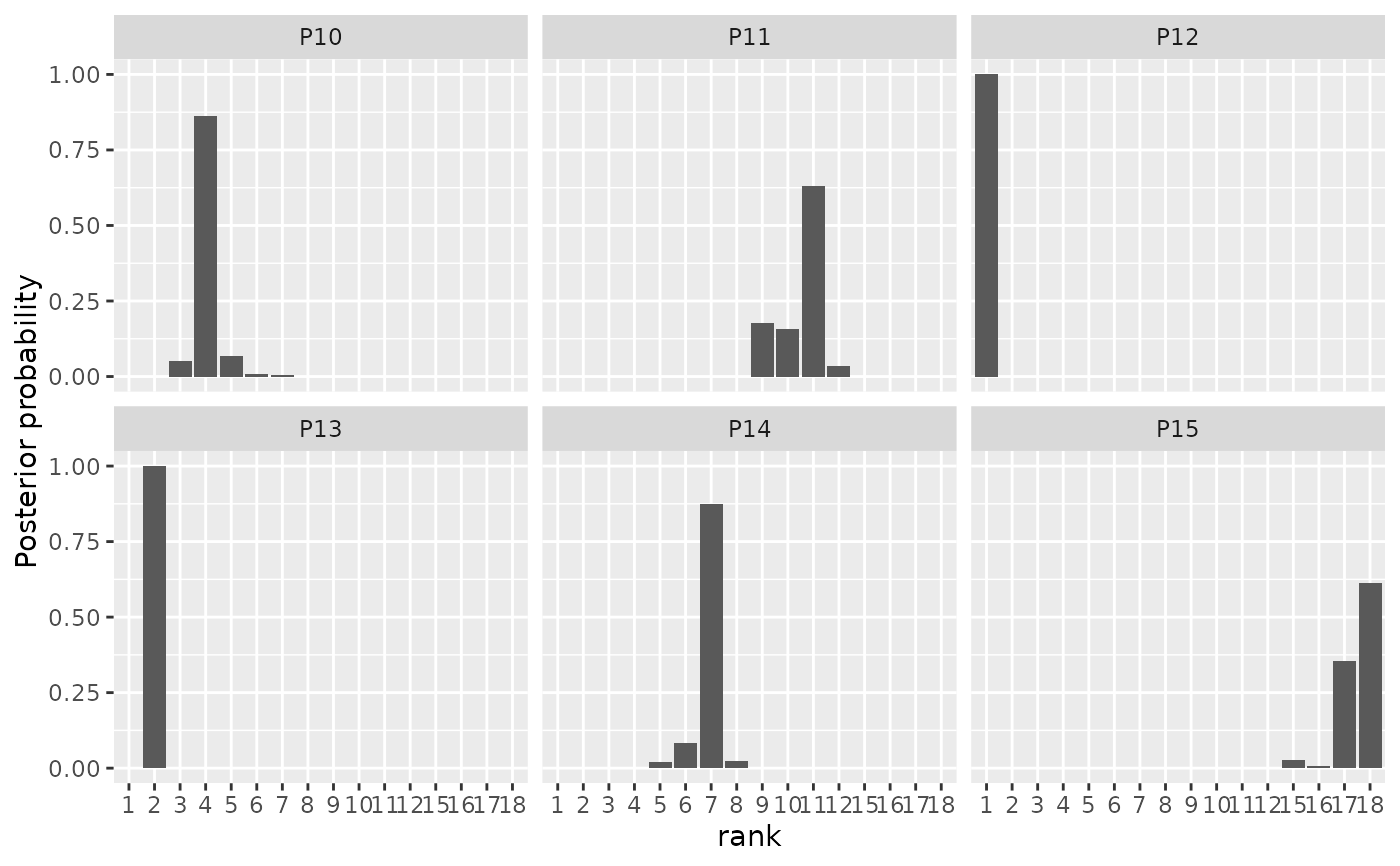 # We can also compute the CP consensus posterior ranking
compute_consensus(model_fit, type = "CP")
#> cluster ranking item cumprob
#> 1 Cluster 1 1 P12 1.000
#> 2 Cluster 1 2 P13 1.000
#> 3 Cluster 1 3 P9 0.876
#> 4 Cluster 1 4 P10 0.916
#> 5 Cluster 1 5 P17 0.748
#> 6 Cluster 1 6 P7 0.882
#> 7 Cluster 1 7 P14 0.977
#> 8 Cluster 1 8 P16 0.845
#> 9 Cluster 1 9 P5 0.605
#> 10 Cluster 1 10 P1 0.743
#> 11 Cluster 1 11 P11 0.965
#> 12 Cluster 1 12 P19 1.000
#> 13 Cluster 1 13 P20 0.582
#> 14 Cluster 1 14 P18 1.000
#> 15 Cluster 1 15 P6 0.916
#> 16 Cluster 1 16 P4 0.727
#> 17 Cluster 1 17 P2 0.780
#> 18 Cluster 1 18 P15 1.000
#> 19 Cluster 1 19 P3 1.000
#> 20 Cluster 1 20 P8 1.000
# And we can compute the posterior intervals:
# First we compute the interval for alpha
compute_posterior_intervals(model_fit, parameter = "alpha")
#> parameter mean median hpdi central_interval
#> 1 alpha 10.632 10.704 [8.468,12.259] [8.473,12.352]
# Then we compute the interval for all the items
compute_posterior_intervals(model_fit, parameter = "rho")
#> parameter item mean median hpdi central_interval
#> 1 rho P1 10 10 [9,12] [9,12]
#> 2 rho P2 17 17 [16,18] [16,18]
#> 3 rho P3 19 19 [19] [19]
#> 4 rho P4 16 16 [16,18] [15,18]
#> 5 rho P5 9 9 [3][6,11] [3,11]
#> 6 rho P6 15 15 [15,16] [15,18]
#> 7 rho P7 6 6 [5,7] [5,7]
#> 8 rho P8 20 20 [20] [20]
#> 9 rho P9 3 3 [3,4] [3,4]
#> 10 rho P10 4 4 [3,5] [3,5]
#> 11 rho P11 11 11 [9,11] [9,12]
#> 12 rho P12 1 1 [1] [1]
#> 13 rho P13 2 2 [2] [2]
#> 14 rho P14 7 7 [6,7] [6,7]
#> 15 rho P15 18 18 [17,18] [15,18]
#> 16 rho P16 8 8 [8,9] [8,9]
#> 17 rho P17 5 5 [5,6][8] [5,8]
#> 18 rho P18 14 14 [13,14] [13,14]
#> 19 rho P19 12 12 [10][12] [10,12]
#> 20 rho P20 13 13 [13,14] [13,14]
# ANALYSIS OF PAIRWISE PREFERENCES
# The example dataset beach_preferences contains pairwise
# preferences between beaches stated by 60 assessors. There
# is a total of 15 beaches in the dataset.
beach_data <- setup_rank_data(
preferences = beach_preferences
)
# We then run the Bayesian Mallows rank model
# We save the augmented data for diagnostics purposes.
model_fit <- compute_mallows(
data = beach_data,
compute_options = set_compute_options(save_aug = TRUE),
progress_report = set_progress_report(verbose = TRUE))
#> First 1000 iterations of Metropolis-Hastings algorithm completed.
# We can assess the convergence of the scale parameter
assess_convergence(model_fit)
# We can also compute the CP consensus posterior ranking
compute_consensus(model_fit, type = "CP")
#> cluster ranking item cumprob
#> 1 Cluster 1 1 P12 1.000
#> 2 Cluster 1 2 P13 1.000
#> 3 Cluster 1 3 P9 0.876
#> 4 Cluster 1 4 P10 0.916
#> 5 Cluster 1 5 P17 0.748
#> 6 Cluster 1 6 P7 0.882
#> 7 Cluster 1 7 P14 0.977
#> 8 Cluster 1 8 P16 0.845
#> 9 Cluster 1 9 P5 0.605
#> 10 Cluster 1 10 P1 0.743
#> 11 Cluster 1 11 P11 0.965
#> 12 Cluster 1 12 P19 1.000
#> 13 Cluster 1 13 P20 0.582
#> 14 Cluster 1 14 P18 1.000
#> 15 Cluster 1 15 P6 0.916
#> 16 Cluster 1 16 P4 0.727
#> 17 Cluster 1 17 P2 0.780
#> 18 Cluster 1 18 P15 1.000
#> 19 Cluster 1 19 P3 1.000
#> 20 Cluster 1 20 P8 1.000
# And we can compute the posterior intervals:
# First we compute the interval for alpha
compute_posterior_intervals(model_fit, parameter = "alpha")
#> parameter mean median hpdi central_interval
#> 1 alpha 10.632 10.704 [8.468,12.259] [8.473,12.352]
# Then we compute the interval for all the items
compute_posterior_intervals(model_fit, parameter = "rho")
#> parameter item mean median hpdi central_interval
#> 1 rho P1 10 10 [9,12] [9,12]
#> 2 rho P2 17 17 [16,18] [16,18]
#> 3 rho P3 19 19 [19] [19]
#> 4 rho P4 16 16 [16,18] [15,18]
#> 5 rho P5 9 9 [3][6,11] [3,11]
#> 6 rho P6 15 15 [15,16] [15,18]
#> 7 rho P7 6 6 [5,7] [5,7]
#> 8 rho P8 20 20 [20] [20]
#> 9 rho P9 3 3 [3,4] [3,4]
#> 10 rho P10 4 4 [3,5] [3,5]
#> 11 rho P11 11 11 [9,11] [9,12]
#> 12 rho P12 1 1 [1] [1]
#> 13 rho P13 2 2 [2] [2]
#> 14 rho P14 7 7 [6,7] [6,7]
#> 15 rho P15 18 18 [17,18] [15,18]
#> 16 rho P16 8 8 [8,9] [8,9]
#> 17 rho P17 5 5 [5,6][8] [5,8]
#> 18 rho P18 14 14 [13,14] [13,14]
#> 19 rho P19 12 12 [10][12] [10,12]
#> 20 rho P20 13 13 [13,14] [13,14]
# ANALYSIS OF PAIRWISE PREFERENCES
# The example dataset beach_preferences contains pairwise
# preferences between beaches stated by 60 assessors. There
# is a total of 15 beaches in the dataset.
beach_data <- setup_rank_data(
preferences = beach_preferences
)
# We then run the Bayesian Mallows rank model
# We save the augmented data for diagnostics purposes.
model_fit <- compute_mallows(
data = beach_data,
compute_options = set_compute_options(save_aug = TRUE),
progress_report = set_progress_report(verbose = TRUE))
#> First 1000 iterations of Metropolis-Hastings algorithm completed.
# We can assess the convergence of the scale parameter
assess_convergence(model_fit)
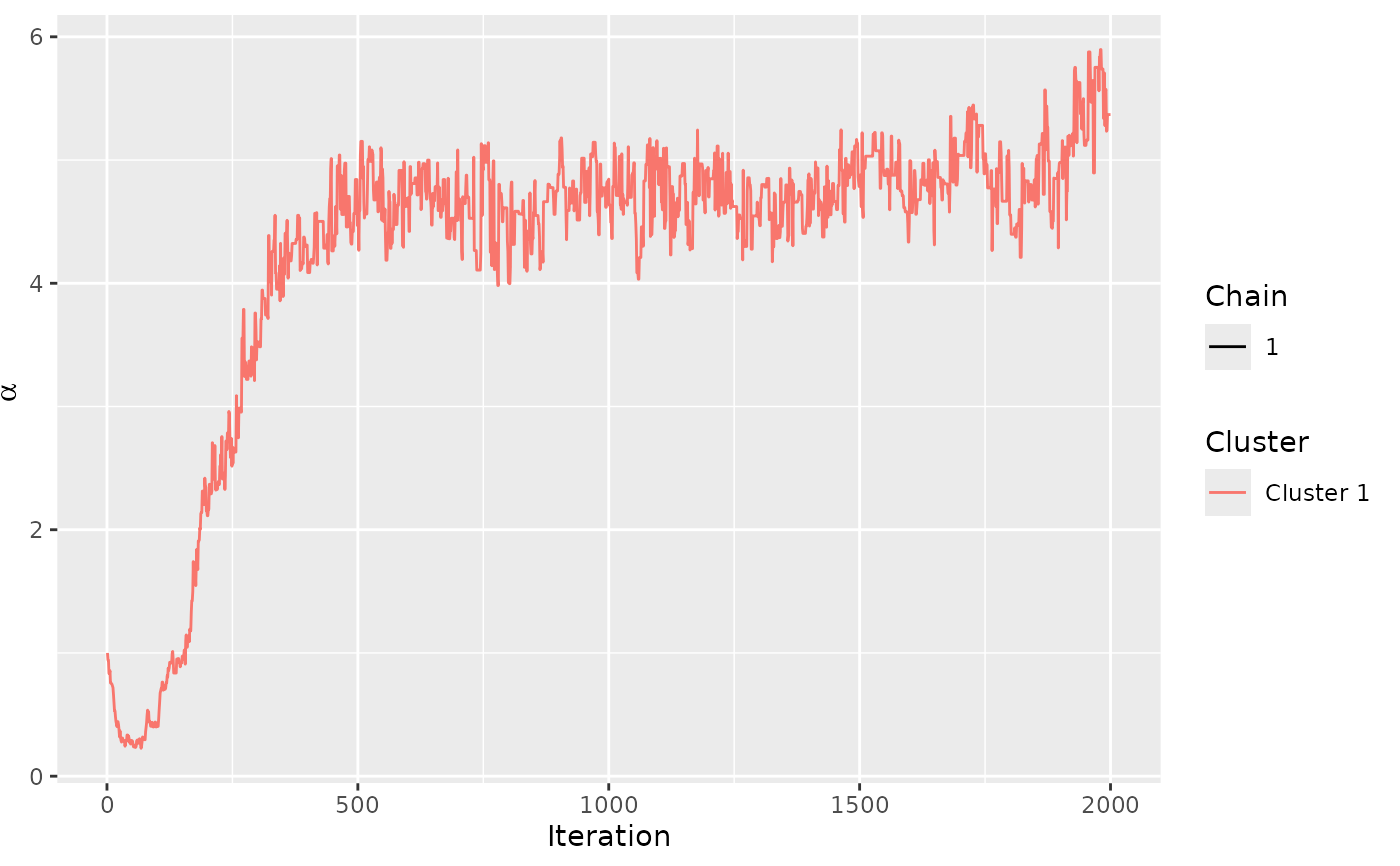 # We can assess the convergence of latent rankings. Here we
# show beaches 1-5.
assess_convergence(model_fit, parameter = "rho", items = 1:5)
# We can assess the convergence of latent rankings. Here we
# show beaches 1-5.
assess_convergence(model_fit, parameter = "rho", items = 1:5)
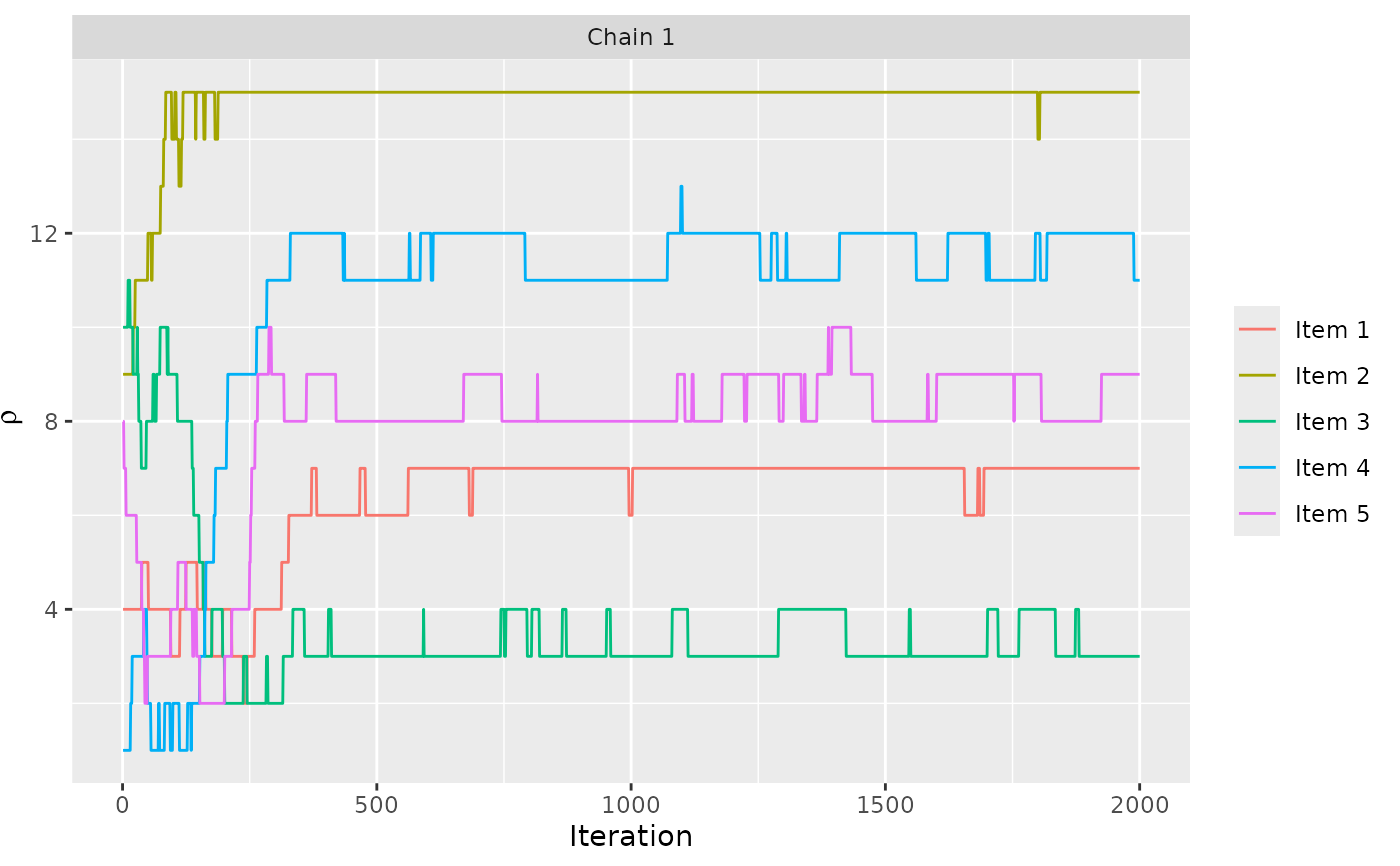 # We can also look at the convergence of the augmented rankings for
# each assessor.
assess_convergence(model_fit, parameter = "Rtilde",
items = c(2, 4), assessors = c(1, 2))
# We can also look at the convergence of the augmented rankings for
# each assessor.
assess_convergence(model_fit, parameter = "Rtilde",
items = c(2, 4), assessors = c(1, 2))
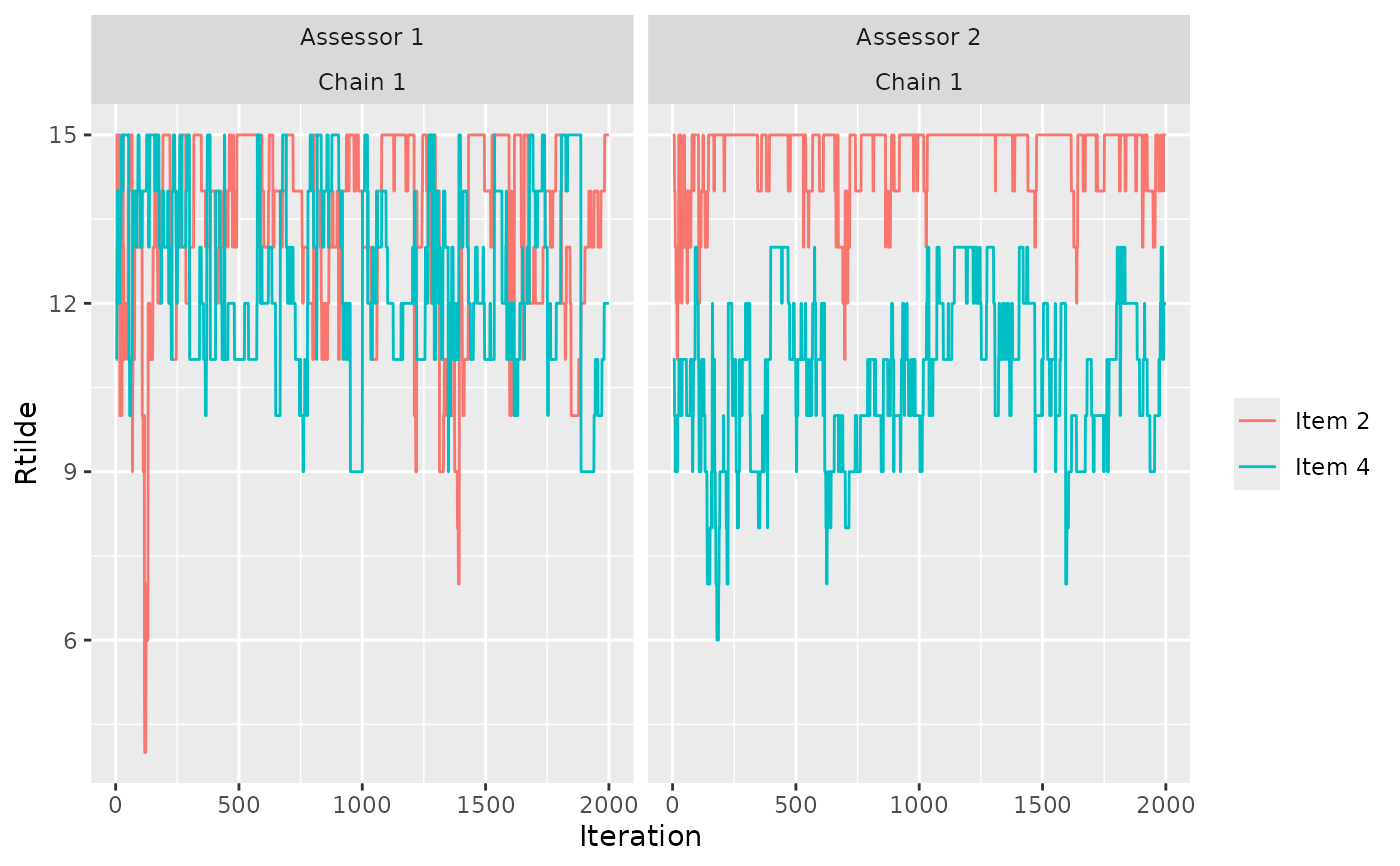 # Notice how, for assessor 1, the lines cross each other, while
# beach 2 consistently has a higher rank value (lower preference) for
# assessor 2. We can see why by looking at the implied orderings in
# beach_tc
subset(get_transitive_closure(beach_data), assessor %in% c(1, 2) &
bottom_item %in% c(2, 4) & top_item %in% c(2, 4))
#> assessor bottom_item top_item
#> 49 2 2 4
# Assessor 1 has no implied ordering between beach 2 and beach 4,
# while assessor 2 has the implied ordering that beach 4 is preferred
# to beach 2. This is reflected in the trace plots.
# CLUSTERING OF ASSESSORS WITH SIMILAR PREFERENCES
if (FALSE) { # \dontrun{
# The example dataset sushi_rankings contains 5000 complete
# rankings of 10 types of sushi
# We start with computing a 3-cluster solution
model_fit <- compute_mallows(
data = setup_rank_data(sushi_rankings),
model_options = set_model_options(n_clusters = 3),
compute_options = set_compute_options(nmc = 10000),
progress_report = set_progress_report(verbose = TRUE))
# We then assess convergence of the scale parameter alpha
assess_convergence(model_fit)
# Next, we assess convergence of the cluster probabilities
assess_convergence(model_fit, parameter = "cluster_probs")
# Based on this, we set burnin = 1000
# We now plot the posterior density of the scale parameters alpha in
# each mixture:
burnin(model_fit) <- 1000
plot(model_fit, parameter = "alpha")
# We can also compute the posterior density of the cluster probabilities
plot(model_fit, parameter = "cluster_probs")
# We can also plot the posterior cluster assignment. In this case,
# the assessors are sorted according to their maximum a posteriori cluster estimate.
plot(model_fit, parameter = "cluster_assignment")
# We can also assign each assessor to a cluster
cluster_assignments <- assign_cluster(model_fit, soft = FALSE)
} # }
# DETERMINING THE NUMBER OF CLUSTERS
if (FALSE) { # \dontrun{
# Continuing with the sushi data, we can determine the number of cluster
# Let us look at any number of clusters from 1 to 10
# We use the convenience function compute_mallows_mixtures
n_clusters <- seq(from = 1, to = 10)
models <- compute_mallows_mixtures(
n_clusters = n_clusters,
data = setup_rank_data(rankings = sushi_rankings),
compute_options = set_compute_options(
nmc = 6000, alpha_jump = 10, include_wcd = TRUE)
)
# models is a list in which each element is an object of class BayesMallows,
# returned from compute_mallows
# We can create an elbow plot
burnin(models) <- 1000
plot_elbow(models)
# We then select the number of cluster at a point where this plot has
# an "elbow", e.g., at 6 clusters.
} # }
# SPEEDING UP COMPUTION WITH OBSERVATION FREQUENCIES With a large number of
# assessors taking on a relatively low number of unique rankings, the
# observation_frequency argument allows providing a rankings matrix with the
# unique set of rankings, and the observation_frequency vector giving the number
# of assessors with each ranking. This is illustrated here for the potato_visual
# dataset
#
# assume each row of potato_visual corresponds to between 1 and 5 assessors, as
# given by the observation_frequency vector
if (FALSE) { # \dontrun{
set.seed(1234)
observation_frequency <- sample.int(n = 5, size = nrow(potato_visual), replace = TRUE)
m <- compute_mallows(
setup_rank_data(rankings = potato_visual, observation_frequency = observation_frequency))
# INTRANSITIVE PAIRWISE PREFERENCES
set.seed(1234)
mod <- compute_mallows(
setup_rank_data(preferences = bernoulli_data),
compute_options = set_compute_options(nmc = 5000),
priors = set_priors(kappa = c(1, 10)),
model_options = set_model_options(error_model = "bernoulli")
)
assess_convergence(mod)
assess_convergence(mod, parameter = "theta")
burnin(mod) <- 3000
plot(mod)
plot(mod, parameter = "theta")
} # }
# CHEKING FOR LABEL SWITCHING
if (FALSE) { # \dontrun{
# This example shows how to assess if label switching happens in BayesMallows
# We start by creating a directory in which csv files with individual
# cluster probabilities should be saved in each step of the MCMC algorithm
# NOTE: For computational efficiency, we use much fewer MCMC iterations than one
# would normally do.
dir.create("./test_label_switch")
# Next, we go into this directory
setwd("./test_label_switch/")
# For comparison, we run compute_mallows with and without saving the cluster
# probabilities The purpose of this is to assess the time it takes to save
# the cluster probabilites.
system.time(m <- compute_mallows(
setup_rank_data(rankings = sushi_rankings),
model_options = set_model_options(n_clusters = 3),
compute_options = set_compute_options(nmc = 500, save_ind_clus = FALSE)))
# With this options, compute_mallows will save cluster_probs2.csv,
# cluster_probs3.csv, ..., cluster_probs[nmc].csv.
system.time(m <- compute_mallows(
setup_rank_data(rankings = sushi_rankings),
model_options = set_model_options(n_clusters = 3),
compute_options = set_compute_options(nmc = 500, save_ind_clus = TRUE)))
# Next, we check convergence of alpha
assess_convergence(m)
# We set the burnin to 200
burnin <- 200
# Find all files that were saved. Note that the first file saved is
# cluster_probs2.csv
cluster_files <- list.files(pattern = "cluster\\_probs[[:digit:]]+\\.csv")
# Check the size of the files that were saved.
paste(sum(do.call(file.size, list(cluster_files))) * 1e-6, "MB")
# Find the iteration each file corresponds to, by extracting its number
iteration_number <- as.integer(
regmatches(x = cluster_files,m = regexpr(pattern = "[0-9]+", cluster_files)
))
# Remove all files before burnin
file.remove(cluster_files[iteration_number <= burnin])
# Update the vector of files, after the deletion
cluster_files <- list.files(pattern = "cluster\\_probs[[:digit:]]+\\.csv")
# Create 3d array, with dimensions (iterations, assessors, clusters)
prob_array <- array(
dim = c(length(cluster_files), m$data$n_assessors, m$n_clusters))
# Read each file, adding to the right element of the array
for(i in seq_along(cluster_files)){
prob_array[i, , ] <- as.matrix(
read.csv(cluster_files[[i]], header = FALSE))
}
# Create an integer array of latent allocations, as this is required by
# label.switching
z <- subset(m$cluster_assignment, iteration > burnin)
z$value <- as.integer(gsub("Cluster ", "", z$value))
z$chain <- NULL
z <- reshape(z, direction = "wide", idvar = "iteration", timevar = "assessor")
z$iteration <- NULL
z <- as.matrix(z)
# Now apply Stephen's algorithm
library(label.switching)
switch_check <- label.switching("STEPHENS", z = z,
K = m$n_clusters, p = prob_array)
# Check the proportion of cluster assignments that were switched
mean(apply(switch_check$permutations$STEPHENS, 1, function(x) {
!all(x == seq(1, m$n_clusters, by = 1))
}))
# Remove the rest of the csv files
file.remove(cluster_files)
# Move up one directory
setwd("..")
# Remove the directory in which the csv files were saved
file.remove("./test_label_switch/")
} # }
# Notice how, for assessor 1, the lines cross each other, while
# beach 2 consistently has a higher rank value (lower preference) for
# assessor 2. We can see why by looking at the implied orderings in
# beach_tc
subset(get_transitive_closure(beach_data), assessor %in% c(1, 2) &
bottom_item %in% c(2, 4) & top_item %in% c(2, 4))
#> assessor bottom_item top_item
#> 49 2 2 4
# Assessor 1 has no implied ordering between beach 2 and beach 4,
# while assessor 2 has the implied ordering that beach 4 is preferred
# to beach 2. This is reflected in the trace plots.
# CLUSTERING OF ASSESSORS WITH SIMILAR PREFERENCES
if (FALSE) { # \dontrun{
# The example dataset sushi_rankings contains 5000 complete
# rankings of 10 types of sushi
# We start with computing a 3-cluster solution
model_fit <- compute_mallows(
data = setup_rank_data(sushi_rankings),
model_options = set_model_options(n_clusters = 3),
compute_options = set_compute_options(nmc = 10000),
progress_report = set_progress_report(verbose = TRUE))
# We then assess convergence of the scale parameter alpha
assess_convergence(model_fit)
# Next, we assess convergence of the cluster probabilities
assess_convergence(model_fit, parameter = "cluster_probs")
# Based on this, we set burnin = 1000
# We now plot the posterior density of the scale parameters alpha in
# each mixture:
burnin(model_fit) <- 1000
plot(model_fit, parameter = "alpha")
# We can also compute the posterior density of the cluster probabilities
plot(model_fit, parameter = "cluster_probs")
# We can also plot the posterior cluster assignment. In this case,
# the assessors are sorted according to their maximum a posteriori cluster estimate.
plot(model_fit, parameter = "cluster_assignment")
# We can also assign each assessor to a cluster
cluster_assignments <- assign_cluster(model_fit, soft = FALSE)
} # }
# DETERMINING THE NUMBER OF CLUSTERS
if (FALSE) { # \dontrun{
# Continuing with the sushi data, we can determine the number of cluster
# Let us look at any number of clusters from 1 to 10
# We use the convenience function compute_mallows_mixtures
n_clusters <- seq(from = 1, to = 10)
models <- compute_mallows_mixtures(
n_clusters = n_clusters,
data = setup_rank_data(rankings = sushi_rankings),
compute_options = set_compute_options(
nmc = 6000, alpha_jump = 10, include_wcd = TRUE)
)
# models is a list in which each element is an object of class BayesMallows,
# returned from compute_mallows
# We can create an elbow plot
burnin(models) <- 1000
plot_elbow(models)
# We then select the number of cluster at a point where this plot has
# an "elbow", e.g., at 6 clusters.
} # }
# SPEEDING UP COMPUTION WITH OBSERVATION FREQUENCIES With a large number of
# assessors taking on a relatively low number of unique rankings, the
# observation_frequency argument allows providing a rankings matrix with the
# unique set of rankings, and the observation_frequency vector giving the number
# of assessors with each ranking. This is illustrated here for the potato_visual
# dataset
#
# assume each row of potato_visual corresponds to between 1 and 5 assessors, as
# given by the observation_frequency vector
if (FALSE) { # \dontrun{
set.seed(1234)
observation_frequency <- sample.int(n = 5, size = nrow(potato_visual), replace = TRUE)
m <- compute_mallows(
setup_rank_data(rankings = potato_visual, observation_frequency = observation_frequency))
# INTRANSITIVE PAIRWISE PREFERENCES
set.seed(1234)
mod <- compute_mallows(
setup_rank_data(preferences = bernoulli_data),
compute_options = set_compute_options(nmc = 5000),
priors = set_priors(kappa = c(1, 10)),
model_options = set_model_options(error_model = "bernoulli")
)
assess_convergence(mod)
assess_convergence(mod, parameter = "theta")
burnin(mod) <- 3000
plot(mod)
plot(mod, parameter = "theta")
} # }
# CHEKING FOR LABEL SWITCHING
if (FALSE) { # \dontrun{
# This example shows how to assess if label switching happens in BayesMallows
# We start by creating a directory in which csv files with individual
# cluster probabilities should be saved in each step of the MCMC algorithm
# NOTE: For computational efficiency, we use much fewer MCMC iterations than one
# would normally do.
dir.create("./test_label_switch")
# Next, we go into this directory
setwd("./test_label_switch/")
# For comparison, we run compute_mallows with and without saving the cluster
# probabilities The purpose of this is to assess the time it takes to save
# the cluster probabilites.
system.time(m <- compute_mallows(
setup_rank_data(rankings = sushi_rankings),
model_options = set_model_options(n_clusters = 3),
compute_options = set_compute_options(nmc = 500, save_ind_clus = FALSE)))
# With this options, compute_mallows will save cluster_probs2.csv,
# cluster_probs3.csv, ..., cluster_probs[nmc].csv.
system.time(m <- compute_mallows(
setup_rank_data(rankings = sushi_rankings),
model_options = set_model_options(n_clusters = 3),
compute_options = set_compute_options(nmc = 500, save_ind_clus = TRUE)))
# Next, we check convergence of alpha
assess_convergence(m)
# We set the burnin to 200
burnin <- 200
# Find all files that were saved. Note that the first file saved is
# cluster_probs2.csv
cluster_files <- list.files(pattern = "cluster\\_probs[[:digit:]]+\\.csv")
# Check the size of the files that were saved.
paste(sum(do.call(file.size, list(cluster_files))) * 1e-6, "MB")
# Find the iteration each file corresponds to, by extracting its number
iteration_number <- as.integer(
regmatches(x = cluster_files,m = regexpr(pattern = "[0-9]+", cluster_files)
))
# Remove all files before burnin
file.remove(cluster_files[iteration_number <= burnin])
# Update the vector of files, after the deletion
cluster_files <- list.files(pattern = "cluster\\_probs[[:digit:]]+\\.csv")
# Create 3d array, with dimensions (iterations, assessors, clusters)
prob_array <- array(
dim = c(length(cluster_files), m$data$n_assessors, m$n_clusters))
# Read each file, adding to the right element of the array
for(i in seq_along(cluster_files)){
prob_array[i, , ] <- as.matrix(
read.csv(cluster_files[[i]], header = FALSE))
}
# Create an integer array of latent allocations, as this is required by
# label.switching
z <- subset(m$cluster_assignment, iteration > burnin)
z$value <- as.integer(gsub("Cluster ", "", z$value))
z$chain <- NULL
z <- reshape(z, direction = "wide", idvar = "iteration", timevar = "assessor")
z$iteration <- NULL
z <- as.matrix(z)
# Now apply Stephen's algorithm
library(label.switching)
switch_check <- label.switching("STEPHENS", z = z,
K = m$n_clusters, p = prob_array)
# Check the proportion of cluster assignments that were switched
mean(apply(switch_check$permutations$STEPHENS, 1, function(x) {
!all(x == seq(1, m$n_clusters, by = 1))
}))
# Remove the rest of the csv files
file.remove(cluster_files)
# Move up one directory
setwd("..")
# Remove the directory in which the csv files were saved
file.remove("./test_label_switch/")
} # }