This vignette contains update syntax for the code examples in Sørensen et al. (2020), since both the underlying code and the user interface are continuously evolving. We refer to Sørensen et al. (2020) for notation and all other details about the models and algorithms.
library(BayesMallows)
set.seed(123)Analysis of complete rankings
We illustrate the case of complete rankings with the potato datasets
described in Section 4 of (Liu et al. 2019). In short, a bag of 20
potatoes was bought, and 12 assessors were asked to rank the potatoes by
weight, first by visual inspection, and next by holding the potatoes in
hand. These datasets are available in BayesMallows as
matrices with names potato_weighing and
potato_visual, respectively. The true ranking of the
potatoes’ weights is available in the vector
potato_true_ranking. In general,
compute_mallows expects ranking datasets to have one row
for each assessor and one column for each item. Each row has to be a
proper permutation, possibly with missing values. We are interested in
the posterior distribution of both the level of agreement between
assessors, as described by
,
and in the latent ranking of the potatoes, as described by
.
We refer to the attached replication script for random number seeds for
exact reproducibility.
We start by defining our data object, which in this case consists of complete rankings.
complete_data <- setup_rank_data(rankings = potato_visual)First, we do a test run to check convergence of the MCMC algorithm,
and then get trace plots with assess_convergence.
bmm_test <- compute_mallows(data = complete_data)
assess_convergence(bmm_test)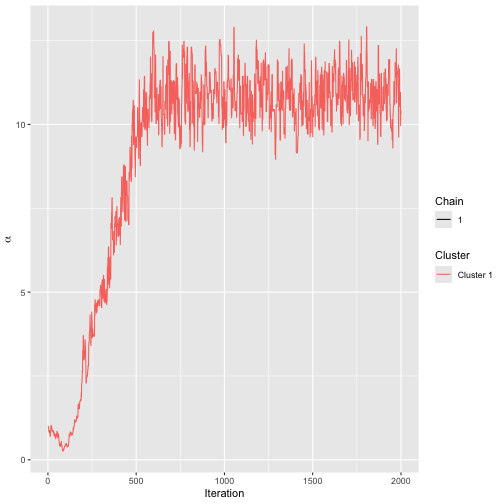
By default, assess_convergence returns a trace plot for
,
shown in the figure above. The algorithm seems to be mixing well after
around 500 iterations. Next, we study the convergence of
.
To avoid overly complex plots, we pick potatoes
by specifying this in the items argument.
assess_convergence(bmm_test, parameter = "rho", items = 1:5)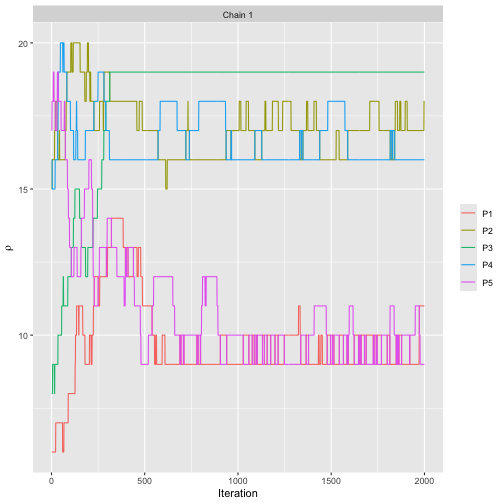
The plot shows that the MCMC algorithm seems to have converged after around 1,000 iterations.
From the trace plots, we decide to discard the first 1,000 MCMC
samples as burn-in. We rerun the algorithm to get 20,000 samples after
burn-in. The object bmm_visual has S3 class
BayesMallows, so we plot the posterior distribution of
with plot.BayesMallows.
bmm_visual <- compute_mallows(
data = complete_data,
compute_options = set_compute_options(nmc = 21000, burnin = 1000)
)
plot(bmm_visual)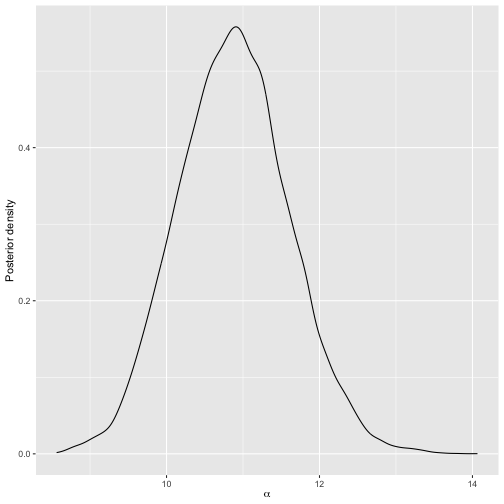
We can also get posterior credible intervals for
using compute_posterior_intervals, which returns both
highest posterior density intervals (HPDI) and central intervals in a
data.frame.
compute_posterior_intervals(bmm_visual, decimals = 1L)
#> parameter mean median hpdi central_interval
#> 1 alpha 10.9 10.9 [9.5,12.3] [9.5,12.3]Next, we can go on to study the posterior distribution of . If the argument is not provided, and the number of items exceeds five, five items are picked at random for plotting. To show all potatoes, we explicitly set .
plot(bmm_visual, parameter = "rho", items = 1:20)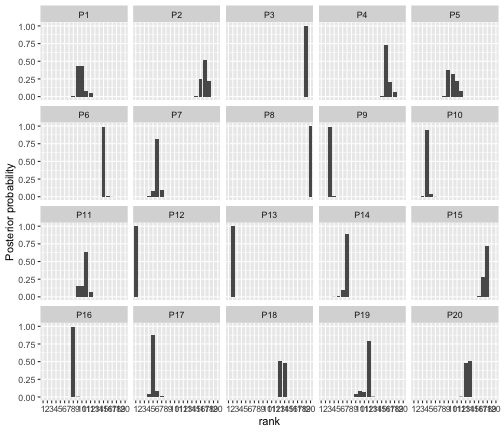
Jumping over the scale parameter
Updating
in every step of the MCMC algorithm may not be necessary, as the number
of posterior samples typically is more than large enough to obtain good
estimates of its posterior distribution. With the
alpha_jump argument, we can tell the MCMC algorithm to
update
only every alpha_jump-th iteration. To update
every 10th time
is updated, we do
bmm_visual <- compute_mallows(
data = complete_data,
compute_options =
set_compute_options(nmc = 21000, burnin = 1000, alpha_jump = 10)
)Other distance metric
By default, compute_mallows uses the footrule distance,
but the user can also choose to use Cayley, Kendall, Hamming, Spearman,
or Ulam distance. Running the same analysis of the potato data with
Spearman distance is done with the command
bmm <- compute_mallows(
data = complete_data,
model_options = set_model_options(metric = "spearman"),
compute_options = set_compute_options(nmc = 21000, burnin = 1000)
)For the particular case of Spearman distance,
BayesMallows only has integer sequences for computing the
exact partition function with 14 or fewer items. In this case a
precomputed importance sampling estimate is part of the package, and
used instead.
Analysis of preference data
Unless the argument error_model to
set_model_options is set, pairwise preference data are
assumed to be consistent within each assessor. These data should be
provided in a dataframe with the following three columns, with one row
per pairwise comparison:
-
assessoris an identifier for the assessor; either a numeric vector containing the assessor index, or a character vector containing the unique name of the assessor. -
bottom_itemis a numeric vector containing the index of the item that was disfavored in each pairwise comparison. -
top_itemis a numeric vector containing the index of the item that was preferred in each pairwise comparison.
A dataframe with this structure can be given in the
preferences argument to setup_rank_data, which
will generate the full set of implied rankings for each assessor as well
as an initial ranking matrix consistent with the pairwise
preferences.
We illustrate with the beach preference data containing stated
pairwise preferences between random subsets of 15 images of beaches, by
60 assessors (Vitelli
et al. 2018). This dataset is provided in the dataframe
beach_preferences, whose first six rows are shown
below:
head(beach_preferences)
#> assessor bottom_item top_item
#> 1 1 2 15
#> 2 1 5 3
#> 3 1 13 3
#> 4 1 4 7
#> 5 1 5 15
#> 6 1 12 6We can define a rank data object based on these preferences.
beach_data <- setup_rank_data(preferences = beach_preferences)It is instructive to compare the computed transitive closure to the stated preferences. Let’s do this for all preferences stated by assessor 1 involving beach 2. We first look at the raw preferences.
subset(beach_preferences, assessor == 1 & (bottom_item == 2 | top_item == 2))
#> assessor bottom_item top_item
#> 1 1 2 15We then use the function get_transitive_closure to
obtain the transitive closure, and then focus on the same subset:
tc <- get_transitive_closure(beach_data)
subset(tc, assessor == 1 & (bottom_item == 2 | top_item == 2))
#> assessor bottom_item top_item
#> 11 1 2 6
#> 44 1 2 15Assessor 1 has performed only one direct comparison involving beach 2, in which the assessor stated that beach 15 is preferred to beach 2. The implied orderings, on the other hand, contain two preferences involving beach 2. In addition to the statement that beach 15 is preferred to beach 2, all the other orderings stated by assessor 1 imply that this assessor prefers beach 6 to beach 2.
Convergence diagnostics
As with the potato data, we can do a test run to assess the
convergence of the MCMC algorithm. This time we use the
beach_data object that we generated above, based on the
stated preferences. We also set save_aug = TRUE to save the
augmented rankings in each MCMC step, hence letting us assess the
convergence of the augmented rankings.
bmm_test <- compute_mallows(
data = beach_data,
compute_options = set_compute_options(save_aug = TRUE))Running assess_convergence for
and
shows good convergence after 1000 iterations.
assess_convergence(bmm_test)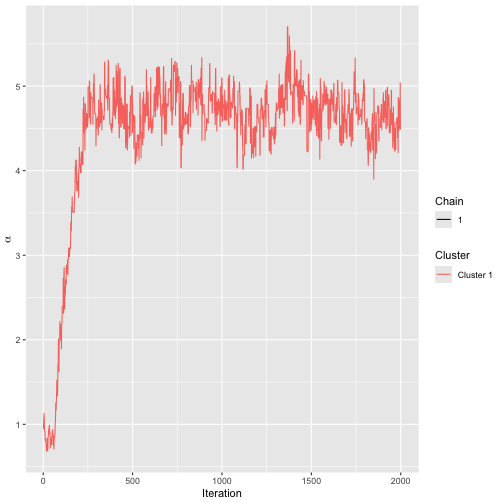
assess_convergence(bmm_test, parameter = "rho", items = 1:6)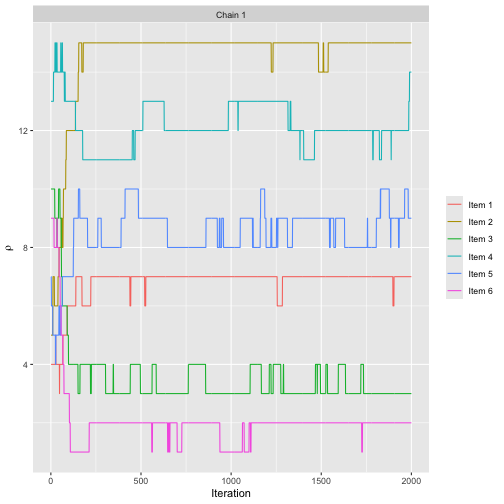
To check the convergence of the data augmentation scheme, we need to
set parameter = "Rtilde", and also specify which items and
assessors to plot. Let us start by considering items 2, 6, and 15 for
assessor 1, which we studied above.
assess_convergence(
bmm_test, parameter = "Rtilde", items = c(2, 6, 15), assessors = 1)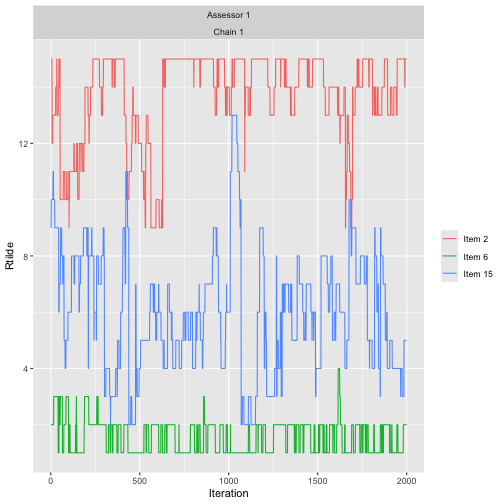
The convergence plot illustrates how the augmented rankings vary, while also obeying their implied ordering.
By further investigation of the transitive closure, we find that no orderings are implied between beach 1 and beach 15 for assessor 2. That is, the following statement returns zero rows.
subset(tc, assessor == 2 & bottom_item %in% c(1, 15) & top_item %in% c(1, 15))
#> [1] assessor bottom_item top_item
#> <0 rows> (or 0-length row.names)With the following command, we create trace plots to confirm this:
assess_convergence(
bmm_test, parameter = "Rtilde", items = c(1, 15), assessors = 2)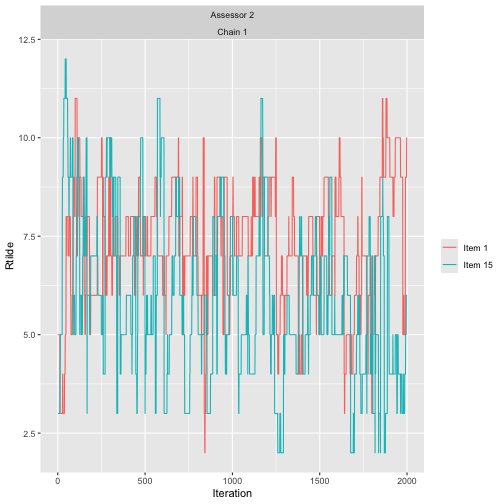
As expected, the traces of the augmented rankings for beach 1 and 15 for assessor 2 do cross each other, since no ordering is implied between them.
Ideally, we should look at trace plots for augmented ranks for more
assessors to be sure that the algorithm is close to convergence. We can
plot assessors 1-8 by setting assessors = 1:8. We also
quite arbitrarily pick items 13-15, but the same procedure can be
repeated for other items.
assess_convergence(
bmm_test, parameter = "Rtilde", items = 13:15, assessors = 1:8)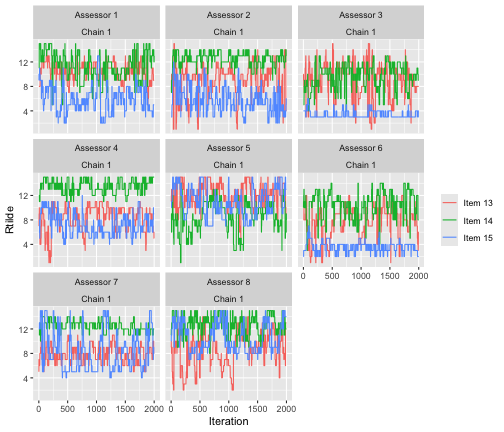
The plot indicates good mixing.
Posterior distributions
Based on the convergence diagnostics, and being fairly conservative, we discard the first 2,000 MCMC iterations as burn-in, and take 20,000 additional samples.
bmm_beaches <- compute_mallows(
data = beach_data,
compute_options =
set_compute_options(nmc = 22000, burnin = 2000, save_aug = TRUE)
)The posterior distributions of
and
can be studied as shown in the previous sections. Posterior intervals
for the latent rankings of each beach are obtained with
compute_posterior_intervals:
compute_posterior_intervals(bmm_beaches, parameter = "rho")
#> parameter item mean median hpdi central_interval
#> 1 rho Item 1 7 7 [7] [7]
#> 2 rho Item 2 15 15 [15] [15]
#> 3 rho Item 3 3 3 [3,4] [3,4]
#> 4 rho Item 4 12 12 [11,13] [11,14]
#> 5 rho Item 5 9 9 [8,10] [8,10]
#> 6 rho Item 6 2 2 [1,2] [1,2]
#> 7 rho Item 7 8 8 [8,9] [8,10]
#> 8 rho Item 8 12 12 [11,13] [11,14]
#> 9 rho Item 9 1 1 [1,2] [1,2]
#> 10 rho Item 10 6 6 [5,6] [5,6]
#> 11 rho Item 11 4 4 [3,4] [3,5]
#> 12 rho Item 12 13 13 [12,14] [12,14]
#> 13 rho Item 13 10 10 [9,10] [9,10]
#> 14 rho Item 14 13 14 [11,14] [11,14]
#> 15 rho Item 15 5 5 [4,5] [4,6]We can also rank the beaches according to their cumulative
probability (CP) consensus (Vitelli et al. 2018) and their
maximum posterior (MAP) rankings. This is done with the function
compute_consensus, and the following call returns the CP
consensus:
compute_consensus(bmm_beaches, type = "CP")
#> cluster ranking item cumprob
#> 1 Cluster 1 1 Item 9 0.89815
#> 2 Cluster 1 2 Item 6 1.00000
#> 3 Cluster 1 3 Item 3 0.72665
#> 4 Cluster 1 4 Item 11 0.95160
#> 5 Cluster 1 5 Item 15 0.95400
#> 6 Cluster 1 6 Item 10 0.97645
#> 7 Cluster 1 7 Item 1 1.00000
#> 8 Cluster 1 8 Item 7 0.62585
#> 9 Cluster 1 9 Item 5 0.85950
#> 10 Cluster 1 10 Item 13 1.00000
#> 11 Cluster 1 11 Item 4 0.46870
#> 12 Cluster 1 12 Item 8 0.84435
#> 13 Cluster 1 13 Item 12 0.61905
#> 14 Cluster 1 14 Item 14 0.99665
#> 15 Cluster 1 15 Item 2 1.00000The column cumprob shows the probability of having the
given rank or lower. Looking at the second row, for example, this means
that beach 6 has probability 1 of having latent rank
.
Next, beach 3 has probability 0.738 of having latent rank
.
This is an example of how the Bayesian framework can be used to not only
rank items, but also to give posterior assessments of the uncertainty of
the rankings. The MAP consensus is obtained similarly, by setting
type = "MAP".
compute_consensus(bmm_beaches, type = "MAP")
#> cluster map_ranking item probability
#> 1 Cluster 1 1 Item 9 0.04955
#> 2 Cluster 1 2 Item 6 0.04955
#> 3 Cluster 1 3 Item 3 0.04955
#> 4 Cluster 1 4 Item 11 0.04955
#> 5 Cluster 1 5 Item 15 0.04955
#> 6 Cluster 1 6 Item 10 0.04955
#> 7 Cluster 1 7 Item 1 0.04955
#> 8 Cluster 1 8 Item 7 0.04955
#> 9 Cluster 1 9 Item 5 0.04955
#> 10 Cluster 1 10 Item 13 0.04955
#> 11 Cluster 1 11 Item 4 0.04955
#> 12 Cluster 1 12 Item 8 0.04955
#> 13 Cluster 1 13 Item 14 0.04955
#> 14 Cluster 1 14 Item 12 0.04955
#> 15 Cluster 1 15 Item 2 0.04955Keeping in mind that the ranking of beaches is based on sparse
pairwise preferences, we can also ask: for beach
,
what is the probability of being ranked
top-
by assessor
,
and what is the probability of having latent rank among the
top-.
The function plot_top_k plots these probabilities. By
default, it sets k = 3, so a heatplot of the probability of
being ranked top-3 is obtained with the call:
plot_top_k(bmm_beaches)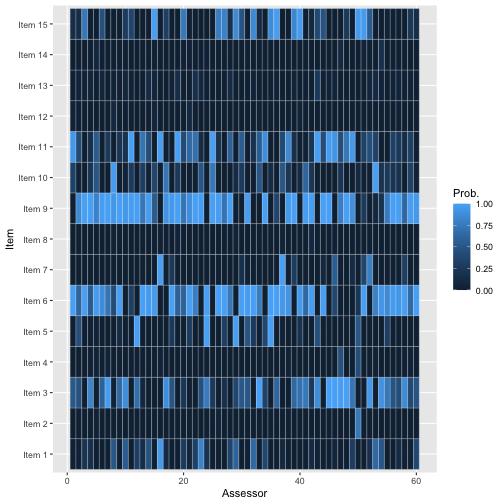
The plot shows, for each beach as indicated on the left axis, the probability that assessor ranks the beach among top-3. For example, we see that assessor 1 has a very low probability of ranking beach 9 among her top-3, while assessor 3 has a very high probability of doing this.
The function predict_top_k returns a dataframe with all
the underlying probabilities. For example, in order to find all the
beaches that are among the top-3 of assessors 1-5 with more than 90 %
probability, we would do:
subset(predict_top_k(bmm_beaches), prob > .9 & assessor %in% 1:5)
#> assessor item prob
#> 301 1 Item 6 0.99435
#> 303 3 Item 6 0.99600
#> 305 5 Item 6 0.97605
#> 483 3 Item 9 1.00000
#> 484 4 Item 9 0.99975
#> 601 1 Item 11 0.95030Note that assessor 2 does not appear in this table, i.e., there are no beaches for which we are at least 90 % certain that the beach is among assessor 2’s top-3.
Clustering
BayesMallows comes with a set of sushi preference data,
in which 5,000 assessors each have ranked a set of 10 types of sushi
(Kamishima
2003). It is interesting to see if we can find subsets of
assessors with similar preferences. The sushi dataset was analyzed with
the BMM by Vitelli et al. (2018), but the results in that paper
differ somewhat from those obtained here, due to a bug in the function
that was used to sample cluster probabilities from the Dirichlet
distribution. We start by defining the data object.
sushi_data <- setup_rank_data(sushi_rankings)Convergence diagnostics
The function compute_mallows_mixtures computes multiple
Mallows models with different numbers of mixture components. It returns
a list of models of class BayesMallowsMixtures, in which
each list element contains a model with a given number of mixture
components. Its arguments are n_clusters, which specifies
the number of mixture components to compute, an optional parameter
cl which can be set to the return value of the
makeCluster function in the parallel package,
and an ellipsis (...) for passing on arguments to
compute_mallows.
Hypothesizing that we may not need more than 10 clusters to find a
useful partitioning of the assessors, we start by doing test runs with
1, 4, 7, and 10 mixture components in order to assess convergence. We
set the number of Monte Carlo samples to 5,000, and since this is a test
run, we do not save within-cluster distances from each MCMC iteration
and hence set include_wcd = FALSE.
library("parallel")
cl <- makeCluster(detectCores())
bmm <- compute_mallows_mixtures(
n_clusters = c(1, 4, 7, 10),
data = sushi_data,
compute_options = set_compute_options(nmc = 5000, include_wcd = FALSE),
cl = cl)
stopCluster(cl)The function assess_convergence automatically creates a
grid plot when given an object of class
BayesMallowsMixtures, so we can check the convergence of
with the command
assess_convergence(bmm)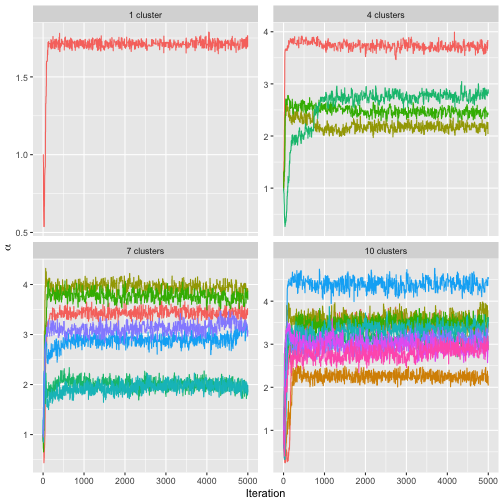
The resulting plot shows that all the chains seem to be close to
convergence quite quickly. We can also make sure that the posterior
distributions of the cluster probabilities
,
have converged properly, by setting
parameter = "cluster_probs".
assess_convergence(bmm, parameter = "cluster_probs")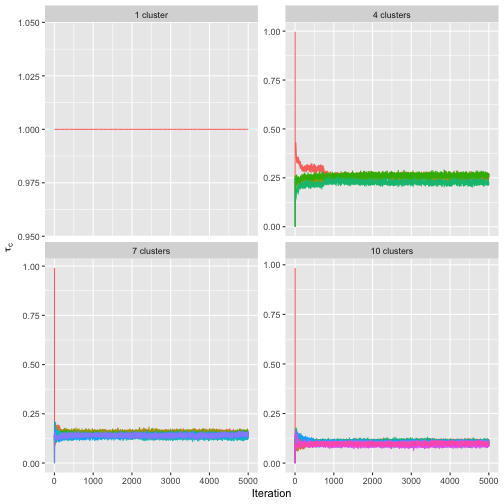
Note that with only one cluster, the cluster probability is fixed at the value 1, while for other number of mixture components, the chains seem to be mixing well.
Deciding on the number of mixture components
Given the convergence assessment of the previous section, we are
fairly confident that a burn-in of 1,000 is sufficient. We run 40,000
additional iterations, and try from 1 to 10 mixture components. Our goal
is now to determine the number of mixture components to use, and in
order to create an elbow plot, we set include_wcd = TRUE to
compute the within-cluster distances in each step of the MCMC algorithm.
Since the posterior distributions of
()
are highly peaked, we save some memory by only saving every 10th value
of
by setting rho_thinning = 10.
cl <- makeCluster(detectCores())
bmm <- compute_mallows_mixtures(
n_clusters = 1:10,
data = sushi_data,
compute_options =
set_compute_options(nmc = 11000, burnin = 1000,
rho_thinning = 10, include_wcd = TRUE),
cl = cl)
stopCluster(cl)We then create an elbow plot:
plot_elbow(bmm)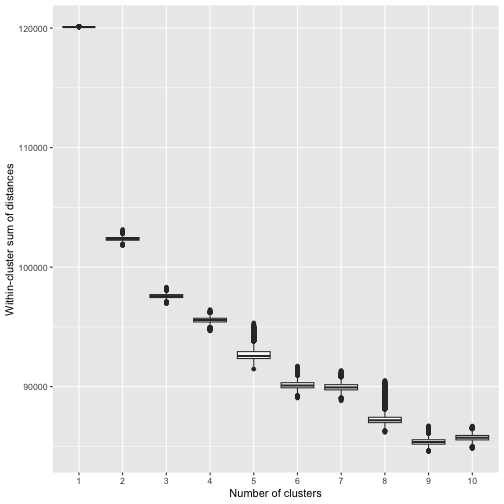
Although not clear-cut, we see that the within-cluster sum of distances levels off at around 5 clusters, and hence we choose to use 5 clusters in our model.
Posterior distributions
Having chosen 5 mixture components, we go on to fit a final model,
still running 10,000 iterations after burnin. This time we call
compute_mallows and set n_clusters = 5. We
also set clus_thinning = 10 to save the cluster assignments
of each assessor in every 10th iteration, and
rho_thinning = 10 to save the estimated latent rank every
10th iteration. Note that thinning is done only for because saving the
values at every iteration would result in very large objects being
stored in memory, thus slowing down computation. For statistical
efficiency, it is best to avoid thinning.
bmm <- compute_mallows(
data = sushi_data,
model_options = set_model_options(n_cluster = 5),
compute_options = set_compute_options(
nmc = 11000, burnin = 1000, clus_thinning = 10, rho_thinning = 10)
)We can plot the posterior distributions of
and
in each cluster using plot.BayesMallows as shown previously
for the potato data.
plot(bmm)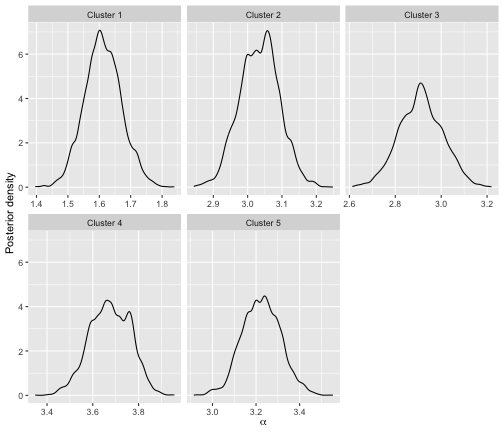
Since there are five clusters, the easiest way of visualizing posterior rankings is by choosing a single item.
plot(bmm, parameter = "rho", items = 1)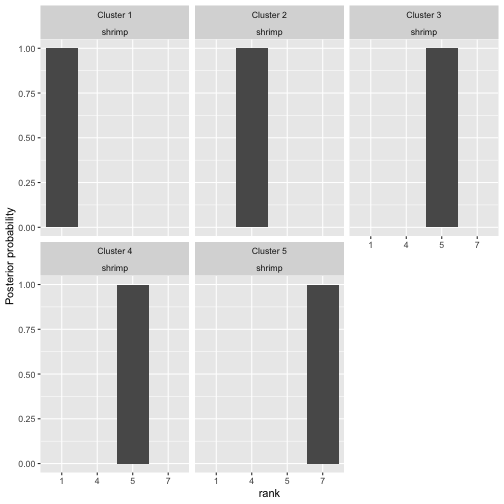
We can also show the posterior distributions of the cluster probabilities.
plot(bmm, parameter = "cluster_probs")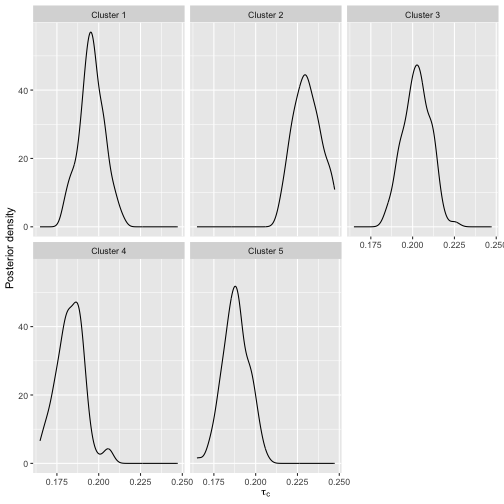
Using the argument parameter = "cluster_assignment", we
can visualize the posterior probability for each assessor of belonging
to each cluster:
plot(bmm, parameter = "cluster_assignment")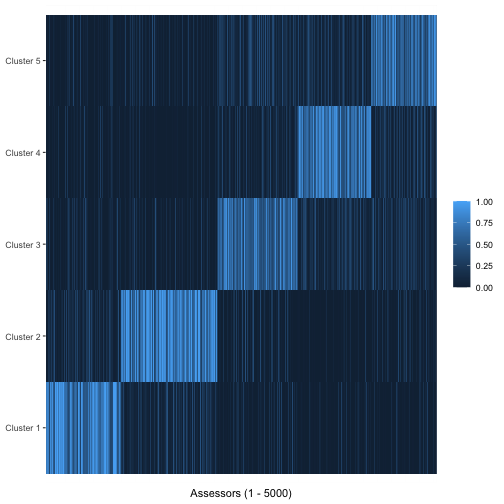
The number underlying the plot can be found using
assign_cluster.
We can find clusterwise consensus rankings using
compute_consensus.
cp_consensus <- compute_consensus(bmm)
reshape(
cp_consensus,
direction = "wide",
idvar = "ranking",
timevar = "cluster",
varying = list(unique(cp_consensus$cluster)),
drop = "cumprob"
)
#> ranking Cluster 1 Cluster 2 Cluster 3 Cluster 4 Cluster 5
#> 1 1 shrimp fatty tuna fatty tuna sea urchin fatty tuna
#> 2 2 sea eel tuna salmon roe fatty tuna sea urchin
#> 3 3 squid sea eel sea urchin salmon roe tuna
#> 4 4 egg shrimp tuna sea eel salmon roe
#> 5 5 fatty tuna tuna roll shrimp shrimp sea eel
#> 6 6 tuna squid tuna roll tuna tuna roll
#> 7 7 tuna roll egg squid squid shrimp
#> 8 8 cucumber roll cucumber roll sea eel tuna roll squid
#> 9 9 salmon roe salmon roe egg egg egg
#> 10 10 sea urchin sea urchin cucumber roll cucumber roll cucumber rollNote that for estimating cluster specific parameters, label switching
is a potential problem that needs to be handled.
BayesMallows ignores label switching issues inside the
MCMC, because it has been shown that this approach is better for
ensuring full convergence of the chain (Jasra, Holmes, and Stephens 2005; Celeux, Hurn, and Robert 2000). MCMC
iterations can be re-ordered after convergence is achieved, for example
by using the implementation of Stephens’ algorithm (Stephens 2000)
provided by the R package label.switching (Papastamoulis
2016). A full example of how to assess label switching is
provided in the examples for the compute_mallows
function.