Example: Single experiment
Example_single.RmdIn the R package, we’ve attached two example datasets from a large drug combination screening experiment on diffuse large B-cell lymphoma. We’ll use these to show some simple use cases of the main functions and how to interpret the results.
Let’s load in the first example and have a look at it
library(bayesynergy)
data("mathews_DLBCL")
y = mathews_DLBCL[[1]][[1]]
x = mathews_DLBCL[[1]][[2]]
head(cbind(y,x))## Viability ibrutinib ispinesib
## [1,] 1.2295618 0.0000 0
## [2,] 1.0376006 0.1954 0
## [3,] 1.1813851 0.7812 0
## [4,] 0.5882688 3.1250 0
## [5,] 0.4666700 12.5000 0
## [6,] 0.2869514 50.0000 0We see that the the measured viability scores are stored in the
vector y, while x is a matrix with two columns
giving the corresponding concentrations where the viability scores were
read off.
Fitting the regression model is simple enough, and can be done on default settings simply by running the following code (where we add the names of the drugs involved, the concentration units for plotting purposes, and calculate the bayes factor).
fit = bayesynergy(y,x, drug_names = c("ibrutinib", "ispinesib"),
units = c("nM","nM"),bayes_factor = T)##
## SAMPLING FOR MODEL 'gp_grid' NOW (CHAIN 1).
## Chain 1:
## Chain 1: Gradient evaluation took 8.8e-05 seconds
## Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0.88 seconds.
## Chain 1: Adjust your expectations accordingly!
## Chain 1:
## Chain 1:
## Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 1: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 1: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 1: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 1: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 1: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 1: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 1: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 1: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 1: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 1:
## Chain 1: Elapsed Time: 2.756 seconds (Warm-up)
## Chain 1: 2.457 seconds (Sampling)
## Chain 1: 5.213 seconds (Total)
## Chain 1:
##
## SAMPLING FOR MODEL 'gp_grid' NOW (CHAIN 2).
## Chain 2:
## Chain 2: Gradient evaluation took 5.2e-05 seconds
## Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 0.52 seconds.
## Chain 2: Adjust your expectations accordingly!
## Chain 2:
## Chain 2:
## Chain 2: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 2: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 2: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 2: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 2: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 2: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 2: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 2: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 2: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 2: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 2: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 2: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 2:
## Chain 2: Elapsed Time: 2.681 seconds (Warm-up)
## Chain 2: 4.878 seconds (Sampling)
## Chain 2: 7.559 seconds (Total)
## Chain 2:
##
## SAMPLING FOR MODEL 'gp_grid' NOW (CHAIN 3).
## Chain 3:
## Chain 3: Gradient evaluation took 5.6e-05 seconds
## Chain 3: 1000 transitions using 10 leapfrog steps per transition would take 0.56 seconds.
## Chain 3: Adjust your expectations accordingly!
## Chain 3:
## Chain 3:
## Chain 3: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 3: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 3: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 3: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 3: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 3: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 3: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 3: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 3: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 3: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 3: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 3: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 3:
## Chain 3: Elapsed Time: 2.257 seconds (Warm-up)
## Chain 3: 2.495 seconds (Sampling)
## Chain 3: 4.752 seconds (Total)
## Chain 3:
##
## SAMPLING FOR MODEL 'gp_grid' NOW (CHAIN 4).
## Chain 4:
## Chain 4: Gradient evaluation took 4.8e-05 seconds
## Chain 4: 1000 transitions using 10 leapfrog steps per transition would take 0.48 seconds.
## Chain 4: Adjust your expectations accordingly!
## Chain 4:
## Chain 4:
## Chain 4: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 4: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 4: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 4: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 4: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 4: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 4: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 4: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 4: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 4: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 4: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 4: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 4:
## Chain 4: Elapsed Time: 2.132 seconds (Warm-up)
## Chain 4: 2.076 seconds (Sampling)
## Chain 4: 4.208 seconds (Total)
## Chain 4:
##
## SAMPLING FOR MODEL 'nointeraction' NOW (CHAIN 1).
## Chain 1:
## Chain 1: Gradient evaluation took 2.6e-05 seconds
## Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0.26 seconds.
## Chain 1: Adjust your expectations accordingly!
## Chain 1:
## Chain 1:
## Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 1: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 1: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 1: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 1: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 1: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 1: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 1: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 1: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 1: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 1:
## Chain 1: Elapsed Time: 0.322 seconds (Warm-up)
## Chain 1: 0.346 seconds (Sampling)
## Chain 1: 0.668 seconds (Total)
## Chain 1:
##
## SAMPLING FOR MODEL 'nointeraction' NOW (CHAIN 2).
## Chain 2:
## Chain 2: Gradient evaluation took 2.7e-05 seconds
## Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 0.27 seconds.
## Chain 2: Adjust your expectations accordingly!
## Chain 2:
## Chain 2:
## Chain 2: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 2: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 2: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 2: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 2: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 2: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 2: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 2: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 2: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 2: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 2: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 2: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 2:
## Chain 2: Elapsed Time: 0.348 seconds (Warm-up)
## Chain 2: 0.198 seconds (Sampling)
## Chain 2: 0.546 seconds (Total)
## Chain 2:
##
## SAMPLING FOR MODEL 'nointeraction' NOW (CHAIN 3).
## Chain 3:
## Chain 3: Gradient evaluation took 1.8e-05 seconds
## Chain 3: 1000 transitions using 10 leapfrog steps per transition would take 0.18 seconds.
## Chain 3: Adjust your expectations accordingly!
## Chain 3:
## Chain 3:
## Chain 3: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 3: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 3: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 3: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 3: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 3: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 3: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 3: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 3: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 3: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 3: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 3: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 3:
## Chain 3: Elapsed Time: 0.332 seconds (Warm-up)
## Chain 3: 0.316 seconds (Sampling)
## Chain 3: 0.648 seconds (Total)
## Chain 3:
##
## SAMPLING FOR MODEL 'nointeraction' NOW (CHAIN 4).
## Chain 4:
## Chain 4: Gradient evaluation took 1.8e-05 seconds
## Chain 4: 1000 transitions using 10 leapfrog steps per transition would take 0.18 seconds.
## Chain 4: Adjust your expectations accordingly!
## Chain 4:
## Chain 4:
## Chain 4: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 4: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 4: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 4: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 4: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 4: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 4: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 4: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 4: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 4: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 4: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 4: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 4:
## Chain 4: Elapsed Time: 0.343 seconds (Warm-up)
## Chain 4: 0.348 seconds (Sampling)
## Chain 4: 0.691 seconds (Total)
## Chain 4:## Calculating the Bayes factorThe resulting model can be summarised by running
summary(fit)## mean se_mean sd 2.5% 50% 97.5% n_eff Rhat
## la_1[1] 0.3301 0.002757 0.0782 1.29e-01 3.44e-01 0.453 804 1.005
## la_2[1] 0.3866 0.002824 0.0652 1.40e-01 3.97e-01 0.458 533 1.008
## log10_ec50_1 0.4866 0.005445 0.1636 2.48e-01 4.48e-01 0.908 902 1.004
## log10_ec50_2 -1.0332 0.019955 1.0356 -3.41e+00 -8.69e-01 0.491 2693 1.000
## slope_1 2.0305 0.020574 0.9436 8.39e-01 1.82e+00 4.462 2103 1.002
## slope_2 1.4765 0.017784 1.0723 1.05e-01 1.22e+00 4.218 3636 1.001
## ell 3.0663 0.034693 1.5244 1.24e+00 2.71e+00 6.958 1931 1.002
## sigma_f 0.8416 0.017847 0.8069 1.70e-01 6.09e-01 2.830 2044 1.002
## s 0.0969 0.000275 0.0149 7.30e-02 9.52e-02 0.131 2956 1.000
## dss_1 33.5129 0.043268 2.8622 2.76e+01 3.36e+01 39.018 4376 1.000
## dss_2 59.4368 0.042907 2.8725 5.36e+01 5.95e+01 64.731 4482 1.000
## rVUS_f 82.7751 0.013496 0.8629 8.11e+01 8.28e+01 84.453 4088 1.000
## rVUS_p0 73.0558 0.031947 2.2230 6.85e+01 7.31e+01 77.257 4842 0.999
## VUS_Delta -9.7193 0.034040 2.3820 -1.46e+01 -9.65e+00 -5.295 4897 1.000
## VUS_syn -9.7633 0.033685 2.3422 -1.46e+01 -9.68e+00 -5.490 4835 1.000
## VUS_ant 0.0440 0.001873 0.1079 5.45e-06 8.02e-05 0.372 3318 1.000
##
## log-Pseudo Marginal Likelihood (LPML) = 50.9839
## Estimated Bayes factor in favor of full model over non-interaction only model: 34.16251which gives posterior summaries of the parameters of the model.
In addition, the model calculates summary statistics of the
monotherapy curves and the dose-response surface including drug
sensitivity scores (DSS) for the two drugs in question, as well as the
volumes that capture the notion of efficacy (rVUS_f),
interaction (VUS_Delta), synergy (VUS_syn) and
interaction (VUS_ant).
As indicated, the total combined drug efficacy is around 80%
(rVUS_f), of which around 70 percentage points can be
attributed to
(rVUS_p0), leaving room for 10 percentage points worth of
synergy (VUS_syn). We can also note that the model is
fairly certain of this effect, with a 95% credible interval given as
(-14.618, -5.49). The certainty of this is also verified by the Bayes
factor, which at 34.16 indicates strong evidence of an interaction
effect present in the model.
Visualization
Monotherapy curves, 2D contour plots
We can also create plots by simply running
plot(fit, plot3D = F)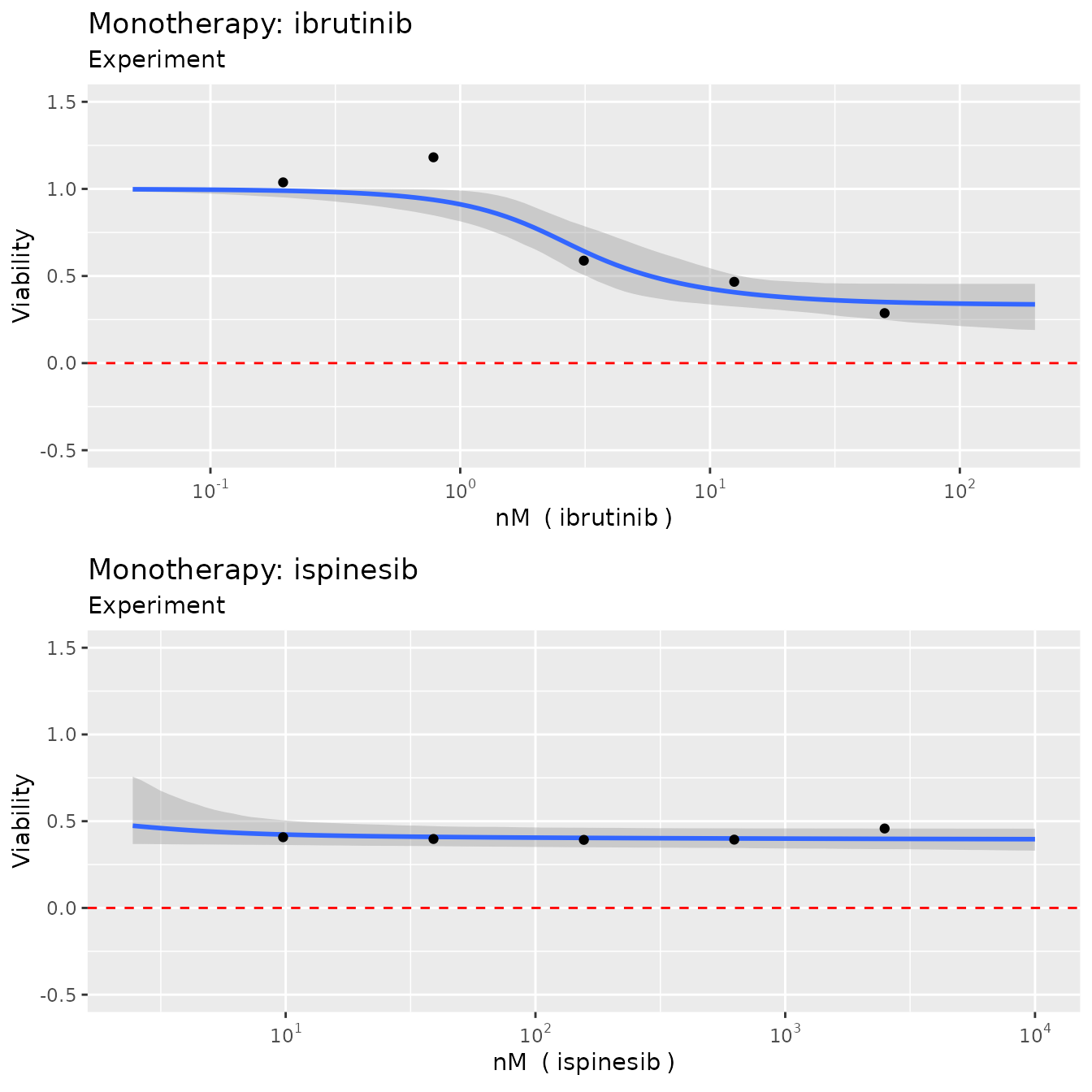

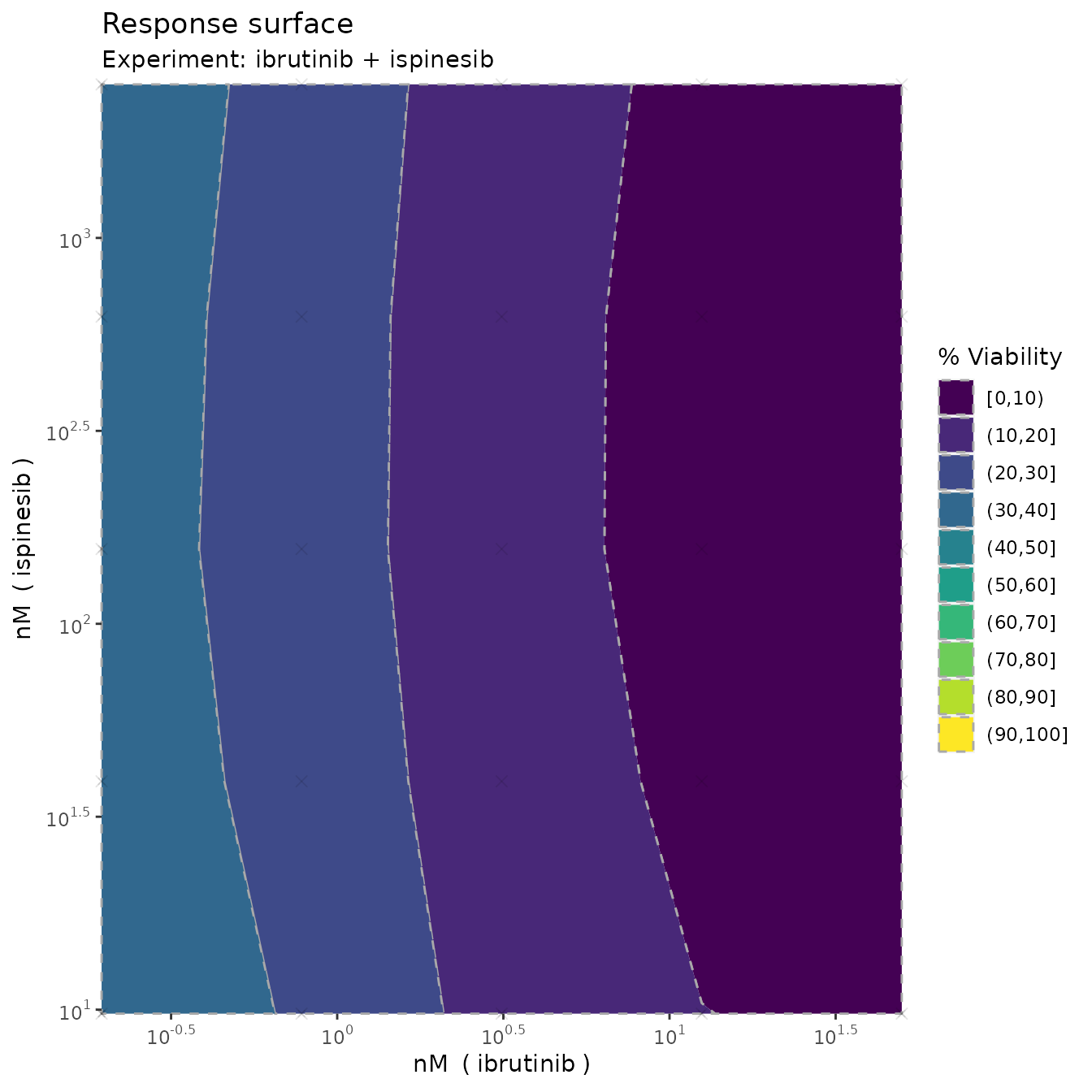
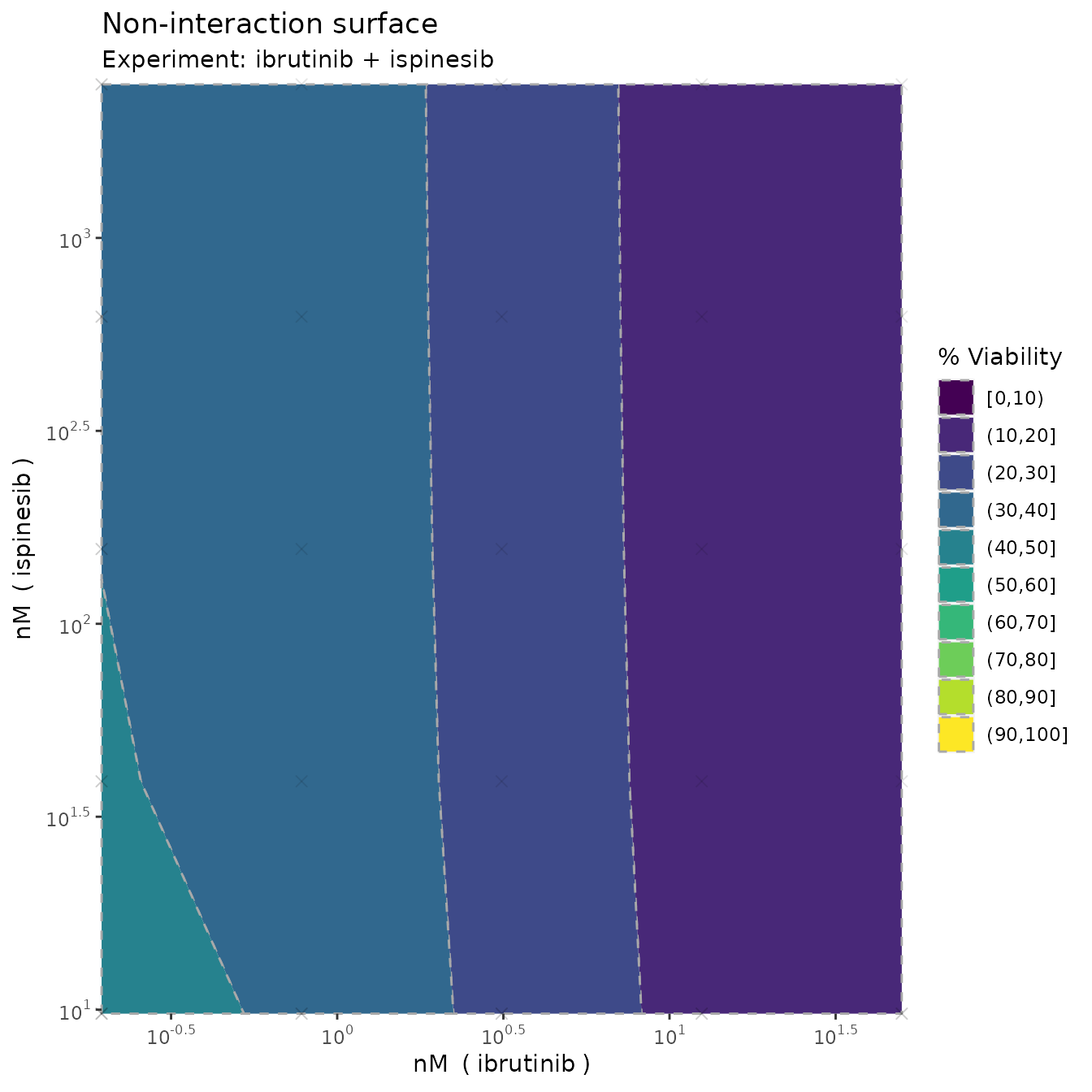

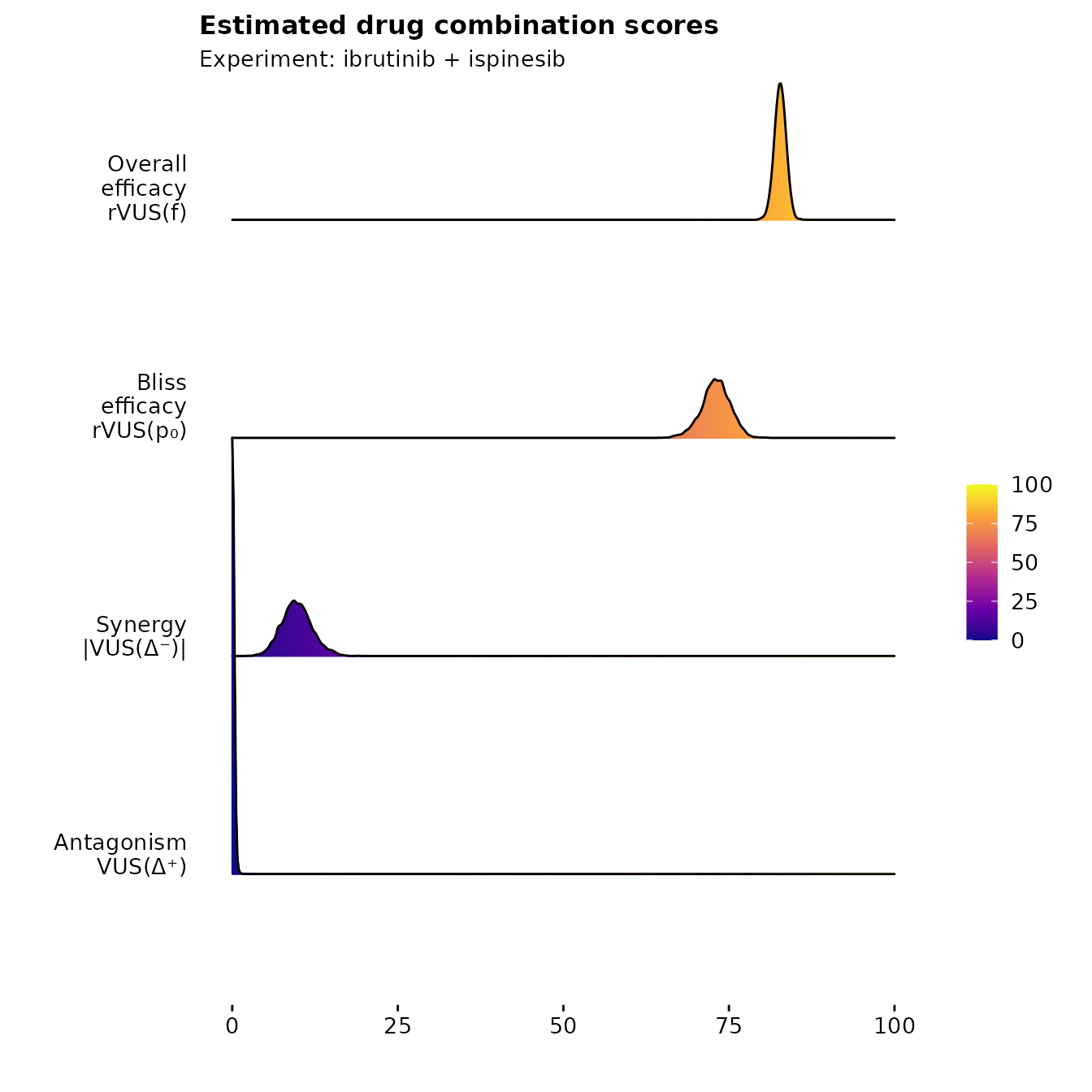
which produces monotherapy curves, monotherapy summary statistics, 2D contour plots of the dose-response function , the non-interaction assumption and the interaction . The last plot displays the scores as discussed previously, with corresponding uncertainty.