Introduction to bayesynergy
bayesynergy.RmdThe bayesynergy R package implements a Bayesian
semi-parametric regression model for estimating the dose-response
function of in-vitro drug combination experiments. The Bayesian
framework offers full uncertainty quantification of the dose response
function and any derived summary statistics, as well as natural handling
of replicates and missing data. The Bayesian model is implemented in RStan, taking
advantage of the efficient ‘No U-Turn Sampler’ as well as variational
Bayes for quick approximations of the true posterior.
The package is further equipped with plotting functions for summarizing a drug response experiment, parallel processing for large drug combination screen, as well as plotting tools for summarizing and comparing these.
Basic usage example
To get started, simply load the package with
We have included an example dataset from a large drug combination screening experiment on diffuse large B-cell lymphoma. To access it, simply run
## Viability ibrutinib ispinesib
## [1,] 1.2295618 0.0000 0
## [2,] 1.0376006 0.1954 0
## [3,] 1.1813851 0.7812 0
## [4,] 0.5882688 3.1250 0
## [5,] 0.4666700 12.5000 0
## [6,] 0.2869514 50.0000 0where y contains the measured post-treatment viability,
and x the corresponding drug concentrations of the two
drugs.
To fit the model, simply run
fit <- bayesynergy(y,x)and wait for the sampling process to complete.
The results can then be plotted to inspect the model fit e.g. for the monotherapies
plot(fit)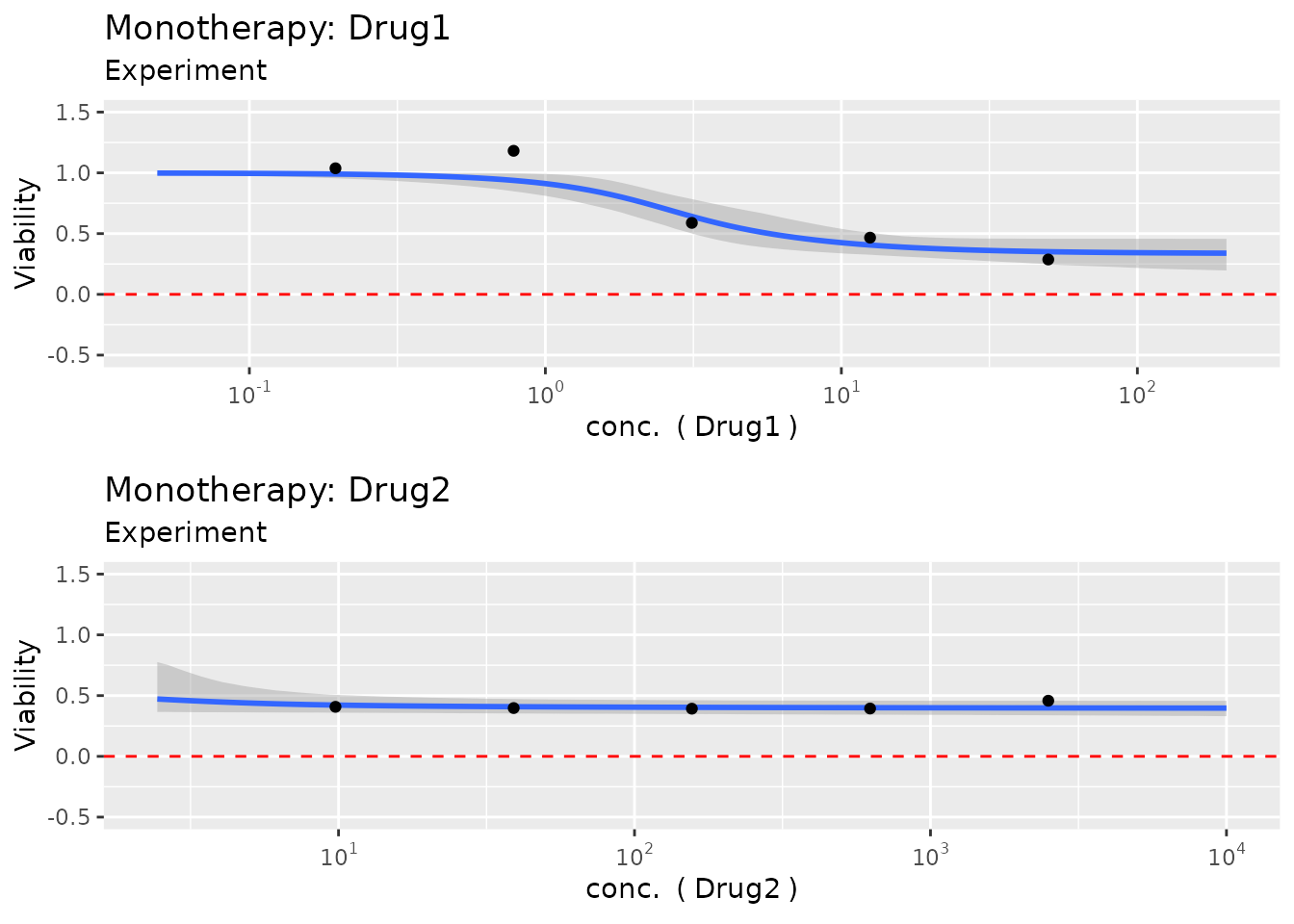
And similarly for the full dose-response surface in an interactive figure
Quantitative measures of drug response and synergy, alongside summaries of the model parameters can be extracted by running
summary(fit)## mean se_mean sd 2.5% 50% 97.5% n_eff Rhat
## la_1[1] 0.3301 0.002757 0.0782 1.29e-01 3.44e-01 0.453 804 1.005
## la_2[1] 0.3866 0.002824 0.0652 1.40e-01 3.97e-01 0.458 533 1.008
## log10_ec50_1 0.4866 0.005445 0.1636 2.48e-01 4.48e-01 0.908 902 1.004
## log10_ec50_2 -1.0332 0.019955 1.0356 -3.41e+00 -8.69e-01 0.491 2693 1.000
## slope_1 2.0305 0.020574 0.9436 8.39e-01 1.82e+00 4.462 2103 1.002
## slope_2 1.4765 0.017784 1.0723 1.05e-01 1.22e+00 4.218 3636 1.001
## ell 3.0663 0.034693 1.5244 1.24e+00 2.71e+00 6.958 1931 1.002
## sigma_f 0.8416 0.017847 0.8069 1.70e-01 6.09e-01 2.830 2044 1.002
## s 0.0969 0.000275 0.0149 7.30e-02 9.52e-02 0.131 2956 1.000
## dss_1 33.5129 0.043268 2.8622 2.76e+01 3.36e+01 39.018 4376 1.000
## dss_2 59.4368 0.042907 2.8725 5.36e+01 5.95e+01 64.731 4482 1.000
## rVUS_f 82.7751 0.013496 0.8629 8.11e+01 8.28e+01 84.453 4088 1.000
## rVUS_p0 73.0558 0.031947 2.2230 6.85e+01 7.31e+01 77.257 4842 0.999
## VUS_Delta -9.7193 0.034040 2.3820 -1.46e+01 -9.65e+00 -5.295 4897 1.000
## VUS_syn -9.7633 0.033685 2.3422 -1.46e+01 -9.68e+00 -5.490 4835 1.000
## VUS_ant 0.0440 0.001873 0.1079 5.45e-06 8.02e-05 0.372 3318 1.000
##
## log-Pseudo Marginal Likelihood (LPML) = 50.9839For a more detailed look into the functionality of the package, please see the other vignettes.